Proximity ligation assay
Proximity ligation assay (in situ PLA) is a technology that extends the capabilities of traditional immunoassays to include direct detection of proteins, protein interactions, extracellular vesicles and post translational modifications with high specificity and sensitivity.[1][2] Protein targets can be readily detected and localized with single molecule resolution and objectively quantified in unmodified cells and tissues. Utilizing only a few cells, sub-cellular events, even transient or weak interactions, are revealed in situ and sub-populations of cells can be differentiated. Within hours, results from conventional co-immunoprecipitation and co-localization techniques can be confirmed.[3]

The PLA principle
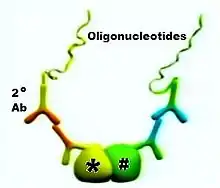
Two primary antibodies raised in different species recognize the target antigen on the proteins of interest (Figure 1). Secondary antibodies (2o Ab) directed against the constant regions of the different primary antibodies, called PLA probes, bind to the primary antibodies (Figure 2).

Each of the PLA probes has a short sequence specific DNA strand attached to it. If the PLA probes are in proximity (that is, if the two original proteins of interest are in proximity, or part of a protein complex, as shown in the figures), the DNA strands can participate in rolling circle DNA synthesis upon addition of two other sequence-specific DNA oligonucleotides together with appropriate substrates and enzymes (Figure 3).
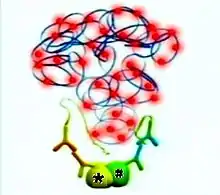
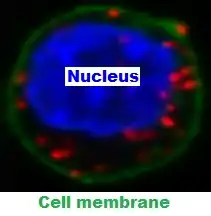
The DNA synthesis reaction results in several-hundredfold amplification of the DNA circle. Next, fluorescent-labeled complementary oligonucleotide probes are added, and they bind to the amplified DNA (Figure 4). The resulting high concentration of fluorescence is easily visible as a distinct bright spot when viewed with a fluorescence microscope.[4] In the specific case shown (Figure 5), the nucleus is enlarged because this is a B-cell lymphoma cell. The two proteins of interest are a B cell receptor and MYD88. The finding of interaction in the cytoplasm was interesting because B cell receptors are thought of as being located in the cell membrane.[5]
Applications
PLA as described above has been used to study aspects of animal development[6][7] and breast cancer[8][9] among many other topics. In situ proximity ligation assays (isPLA) has been applied to antibody validation in human tissues with various advantages over IHC, including increased detection specificity, decreased unspecific staining, and better localization.[10] A variation of the technique (rISH-PLA) has been used to study the association of protein and RNA.[11] Another variation of in situ PLA includes a multiplex PLA assay that makes it possible to visualize multiple protein complexes in parallel.[12] PLA can also be combined with other read out forms such as ELISA,[13] flow cytometry.[14][15] and Western blotting[16]
References
- Löf, Liza; Arngården, Linda; Ebai, Tonge; Landegren, Ulf; Söderberg, Ola; Kamali-Moghaddam, Masood (2017). "Detection of Extracellular Vesicles Using Proximity Ligation Assay with Flow Cytometry Readout—ExoPLA". Current Protocols in Cytometry. 81 (1): 4.8.1–4.8.10. doi:10.1002/cpcy.22. ISSN 1934-9300. PMID 28678418. S2CID 4379414.
- Gullberg M, Gústafsdóttir SM, Schallmeiner E, Jarvius J, Bjarnegård M, Betsholtz C, Landegren U, Fredriksson S (June 2004). "Cytokine detection by antibody-based proximity ligation". Proceedings of the National Academy of Sciences of the United States of America. 101 (22): 8420–4. Bibcode:2004PNAS..101.8420G. doi:10.1073/pnas.0400552101. PMC 420409. PMID 15155907.
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U (December 2006). "Direct observation of individual endogenous protein complexes in situ by proximity ligation". Nature Methods. 3 (12): 995–1000. doi:10.1038/nmeth947. PMID 17072308. S2CID 21819907.
- Gustafsdottir SM, Schallmeiner E, Fredriksson S, Gullberg M, Söderberg O, Jarvius M, Jarvius J, Howell M, Landegren U (October 2005). "Proximity ligation assays for sensitive and specific protein analyses". Analytical Biochemistry. 345 (1): 2–9. doi:10.1016/j.ab.2005.01.018. PMID 15950911.
- Staudt, Louis (February 2017). "Therapy of lymphoma inspired by functional and structural genomics". videocast.nih.gov. National Institutes of Health. Retrieved 3 February 2017.
- Wang S, Yoo S, Kim HY, Wang M, Zheng C, Parkhouse W, Krieger C, Harden N (January 2015). "Detection of in situ protein-protein complexes at the Drosophila larval neuromuscular junction using proximity ligation assay". Journal of Visualized Experiments (95): 52139. doi:10.3791/52139. PMC 4354543. PMID 25650626.
- Kwon J, Jeong SM, Choi I, Kim NH (2016). "ADAM10 Is Involved in Cell Junction Assembly in Early Porcine Embryo Development". PLOS ONE. 11 (4): e0152921. Bibcode:2016PLoSO..1152921K. doi:10.1371/journal.pone.0152921. PMC 4820119. PMID 27043020.
- Karamouzis MV, Dalagiorgou G, Georgopoulou U, Nonni A, Kontos M, Papavassiliou AG (February 2016). "HER-3 targeting alters the dimerization pattern of ErbB protein family members in breast carcinomas". Oncotarget. 7 (5): 5576–97. doi:10.18632/oncotarget.6762. PMC 4868707. PMID 26716646.
- Vincent A, Berthel E, Dacheux E, Magnard C, Venezia NL (April 2016). "BRCA1 affects protein phosphatase 6 signalling through its interaction with ANKRD28". The Biochemical Journal. 473 (7): 949–60. doi:10.1042/BJ20150797. PMID 27026398.
- Lindskog, Cecilia; Backman, Max; Zieba, Agata; Asplund, Anna; Uhlén, Mathias; Landegren, Ulf; Pontén, Fredrik (July 2020). "Proximity Ligation Assay as a Tool for Antibody Validation in Human Tissues". Journal of Histochemistry & Cytochemistry. 68 (7): 515–529. doi:10.1369/0022155420936384. ISSN 0022-1554. PMC 7350078. PMID 32602410.
- Roussis IM, Guille M, Myers FA, Scarlett GP (2016). "RNA Whole-Mount In situ Hybridisation Proximity Ligation Assay (rISH-PLA), an Assay for Detecting RNA-Protein Complexes in Intact Cells". PLOS ONE. 11 (1): e0147967. Bibcode:2016PLoSO..1147967R. doi:10.1371/journal.pone.0147967. PMC 4732756. PMID 26824753.
- Leuchowius KJ, Clausson CM, Grannas K, Erbilgin Y, Botling J, Zieba A, et al. (June 2013). "Parallel visualization of multiple protein complexes in individual cells in tumor tissue". Molecular & Cellular Proteomics. 12 (6): 1563–71. doi:10.1074/mcp.O112.023374. PMC 3675814. PMID 23436906.
- Ebai T, Souza de Oliveira FM, Löf L, Wik L, Schweiger C, Larsson A, et al. (September 2017). "Analytically Sensitive Protein Detection in Microtiter Plates by Proximity Ligation with Rolling Circle Amplification". Clinical Chemistry. 63 (9): 1497–1505. doi:10.1373/clinchem.2017.271833. PMID 28667186.
- Leuchowius KJ, Weibrecht I, Landegren U, Gedda L, Söderberg O (October 2009). "Flow cytometric in situ proximity ligation analyses of protein interactions and post-translational modification of the epidermal growth factor receptor family". Cytometry. Part A. 75 (10): 833–9. doi:10.1002/cyto.a.20771. PMID 19650109. S2CID 2550136.
- Löf L, Arngården L, Olsson-Strömberg U, Siart B, Jansson M, Dahlin JS, et al. (April 2017). "Flow Cytometric Measurement of Blood Cells with BCR-ABL1 Fusion Protein in Chronic Myeloid Leukemia". Scientific Reports. 7 (1): 623. Bibcode:2017NatSR...7..623L. doi:10.1038/s41598-017-00755-y. PMC 5429594. PMID 28377570.
- Liu Y, Gu J, Hagner-McWhirter Å, Sathiyanarayanan P, Gullberg M, Söderberg O, et al. (November 2011). "Western blotting via proximity ligation for high performance protein analysis". Molecular & Cellular Proteomics. 10 (11): O111.011031. doi:10.1074/mcp.O111.011031. PMC 3226413. PMID 21813417.