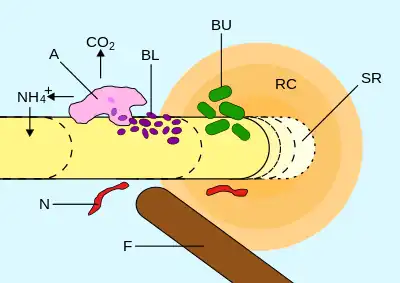
Rhizosphere
The rhizosphere is the narrow region of soil or substrate that is directly influenced by root secretions and associated soil microorganisms known as the root microbiome.[2] Soil pores in the rhizosphere can contain many bacteria and other microorganisms that feed on sloughed-off plant cells, termed rhizodeposition,[3] and the proteins and sugars released by roots, termed root exudates.[4] This symbiosis leads to more complex interactions, influencing plant growth and competition for resources. Much of the nutrient cycling and disease suppression by antibiotics required by plants, occurs immediately adjacent to roots due to root exudates and metabolic products of symbiotic and pathogenic communities of microorganisms.[5] The rhizosphere also provides space to produce allelochemicals to control neighbours and relatives.[6]
The rhizoplane refers to the root surface including its associated soil particles which closely interact with each other.[7] The plant-soil feedback loop and other physical factors occurring at the plant-root soil interface are important selective pressures in communities and growth in the rhizosphere and rhizoplane.[8]
Background
The term "rhizosphere" was used first in 1904 by the German plant physiologist Lorenz Hiltner to describe how plant roots interface with surrounding soil.[9][10] The prefix rhiza- comes from the Greek, and means "root". Hiltner postulated the rhizosphere was a region surrounding the plant roots, and populated with microorganisms under some degree of control by chemicals released from the plant roots.[9][11]
Chemical interactions
| Part of a series on |
| Microbiomes |
|---|
 |
|
Chemical availability
Plant roots may exude 20-40% of the sugars and organic acids - photosynthetically fixed carbon.[12] Plant root exudates, such as organic acids, change the chemical structure and the biological communities of the rhizosphere in comparison with the bulk soil or parent soil. Concentrations of organic acids and saccharides affect the ability of the biological communities to shuttle phosphorus, nitrogen,[13][14] potassium and water to the root cap,[15] and the total availability of iron to the plant and to its neighbors.[16] The ability of the plant's root and its associated soil microorganisms to provide specific transport proteins affect the availability of iron and other minerals for it and its neighbors. This can affect the composition of the community and its fitness.
Root exudates come in the form of chemicals released into the rhizosphere by cells in the roots and cell waste referred to as "rhizodeposition." This rhizodeposition comes in various forms of organic carbon and nitrogen that provide for the communities around plant roots and dramatically affect the chemistry surrounding the roots.[17] Exopolysaccharides, such as polyglycolide (PGA), affect the ability of roots to uptake water by maintaining the physical stability of the soil carbon sponge and control the flow of water.[18] For example, a tomato field study showed that exopolysaccharides extracted from the rhizosphere were different (total sugar amounts and mean infrared measurements) depending on the tomato varieties grown,[19] and that under water deficit conditions (limited irrigation) the increase in exopolysaccharide production and microbial activity affected water retention in the soil and field performance of tomato.[20] In potato cultivar root exudates, phenols and lignins comprise the greatest number of ion influencing compounds regardless of growing location; however, the intensity of different compounds was found to be influenced by soils and environmental conditions, resulting in variation amongst nitrogen compounds, lignins, phenols, carbohydrates, and amines.[21]
Allelochemicals
Chemicals connected to allelopathy: flavonols [22] carbohydrates and application by root hairs[23] phenols[24] Positive allelopathic pathways and definitions of interactions between plant-plant and plant-microbe,[25] positive plant-microbe in the form of systematic resistance[26]
Although it goes beyond the rhizosphere area, it is notable that some plants secrete allelochemicals from their roots which inhibit the growth of other organisms. For example, garlic mustard produces a chemical that is believed to prevent mutualisms forming between the surrounding trees and mycorrhiza in mesic North American temperate forests where it is an invasive species.[27]
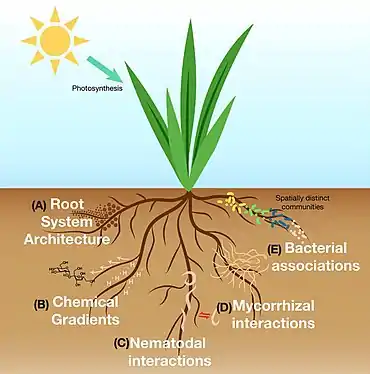
Ecology of the rhizosphere
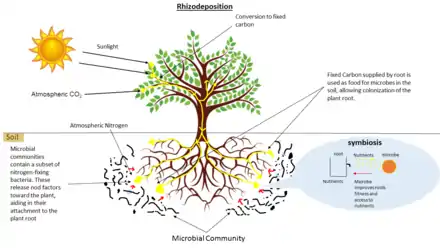
Rhizodeposition allows for the growth of communities of microorganisms directly surrounding and inside plant roots. This leads to complex interactions between species including mutualism, predation/parasitism, and competition.
Predation
Predation is considered to be top-down because these interactions decrease the population, but the closeness of the interactions of species directly affects the availability of resources causing the population to also be affected by bottom-up controls.[31] Without soil fauna, microbes that directly prey upon competitors of plants and plant mutualists, interactions within the rhizosphere would be antagonistic toward the plants. Soil fauna provide the top-down component of the rhizosphere while also allowing for the bottom-up increase in nutrients from rhizodeposition and inorganic nitrogen. The complexity of these interactions has also been shown through experiments with common soil fauna, such as nematodes and protists. Predation by bacterial-feeding nematodes was shown to influence nitrogen availability and plant growth.[32] There was also an increase in the populations of bacteria to which nematodes were added. Predation upon Pseudomonas by amoeba shows predators are able to upregulate toxins produced by prey without direct interaction using supernatant.[33] The ability of predators to control the expression and production of biocontrol agents in prey without direct contact is related to the evolution of prey species to signals of high predator density and nutrient availability.
The food web in the rhizosphere can be considered as three different channels with two different sources of energy: the detritus-dependent channels are fungi and bacterial species, and the root energy-dependent channel consists of nematodes, symbiotic species, and some arthropods.[31] This food web is constantly in flux, since the amount of detritus available and the rate of root sloughing changes as roots grow and age. This bacterial channel is considered to be a faster channel because of the ability of species to focus on more accessible resources in the rhizosphere and have faster regeneration times compared with the fungal channel. All three of these channels are also interrelated to the roots that form the base of the rhizosphere ecosystem and the predators, such as the nematodes and protists, that prey upon many of the same species of microflora.
Competition
Allelopathy and autotoxicity and negative root-root communications[34][16]
The competition between plants due to released exudates is dependent upon geometrical properties, which determine the capacity of interception of exudates from any point in relation to the plants’ roots, and physicochemical properties, which determine the capacity of each root to take up exudates in the area.[35] Geometrical properties are the density of roots, root diameter, and distribution of the roots and physicochemical properties are exudation rate, decay rate of exudates, and the properties of the environment that affect diffusion. These properties define the rhizosphere of roots and the likelihood that plants can directly compete with neighbors.
Plants and soil microflora indirectly compete against one another by tying up limiting resources, such as carbon and nitrogen, into their biomass.[36] This competition can occur at varying rates due to the ratio of carbon to nitrogen in detritus and the ongoing mineralization of nitrogen in the soil. Mycorrhizae and heterotrophic soil microorganisms compete for both carbon and nitrogen depending upon which is limiting at the time, which itself heavily depends on the species, scavenging abilities, and the environmental conditions affecting nitrogen input. Plants are less successful at uptake of organic nitrogen, such as amino acids, than the soil microflora that exist in the rhizosphere.[37] This informs other mutualistic relationships formed by plants with relation to nitrogen uptake.
Competition over other resources, such as oxygen in limited environments, are directly affected by the spatial and temporal locations of species and the rhizosphere. In methanotrophs, proximity to higher density roots and the surface are important and help to determine where they are dominant over heterotrophs in rice paddies.[38]
The weak connection between the various channels of energy is important in regulation in the populations of both predator and prey and the availability of resources to the biome. Strong connections between resource-consumer and consumer-consumer create coupled systems of oscillators which are then determined by the nature of the available resources.[39] These systems can then be considered cyclical, quasi-periodic, or chaotic.
Mutualism
Plants secrete many compounds through their roots to serve symbiotic functions in the rhizosphere. Strigolactones, secreted and detected by mycorrhizal fungi, stimulate the germination of spores and initiate changes in the mycorrhiza that allow it to colonize the root. The parasitic plant, Striga also detects the presence of strigolactones and will germinate when it detects them; they will then move into the root, feeding off the nutrients present.[40][41]
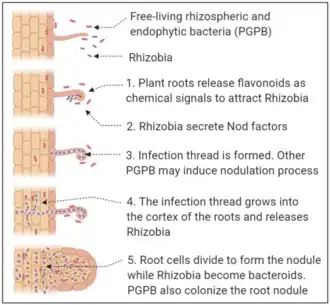
Symbiotic nitrogen-fixing bacteria, such as Rhizobium species, detect compounds like flavonoids secreted by the roots of leguminous plants and then produce nod factors that signal to the plant that they are present and will lead to the formation of root nodules. Bacteria are housed in symbiosomes in these nodules where they are sustained by nutrients from the plant and convert nitrogen gas to a form that can be used by the plant.[42] Non-symbiotic (or "free-living") nitrogen-fixing bacteria may reside in the rhizosphere just outside the roots of certain plants (including many grasses) and similarly "fix" nitrogen gas in the nutrient-rich plant rhizosphere. Even though these organisms are thought to be only loosely associated with plants they inhabit, they may respond very strongly to the status of the plants. For example, nitrogen-fixing bacteria in the rhizosphere of the rice plant exhibit diurnal cycles that mimic plant behavior and tend to supply more fixed nitrogen during growth stages when the plant exhibits a high demand for nitrogen.[43]
In exchange for the resources and shelter that plants and roots provide, fungi and bacteria control pathogenic microbes.[44] The fungi that perform such activities also serve close relationships with species of plants in the form of mycorrhizal fungi, which are diverse in how they relate to plants. Arbuscular mycorrhizal fungi and the bacteria that make the rhizosphere their home also form close relationships in order to be more competitive.[45] which plays into the bigger cycles of nutrients that impact the ecosystem, such as biogeochemical pathways.[16]
Community structure
The rhizosphere has been referred to as an information super highway because of the proximity of data points, which include roots, and organisms in the soil, and the methods for transferring data using exudates and communities.[34] This description has been used to explain the complex interactions that plants, their fungal mutualists, and the bacterial species that live in the rhizosphere have entered into over the course of their evolution. Certain species like Trichoderma are interesting because of their ability to select for species in this complex web. Trichoderma is a biological control agent because of evidence that it can reduce plant pathogens in the rhizosphere.[46] Plants themselves also affect which bacterial species in the rhizosphere are selected against because of the introduction of exudates and the relationships that it maintains. The control of which species are in these small diversity hotspots can drastically affect the capacity of these spaces and future conditions for future ecologies.[44][6]
Microbial consortium
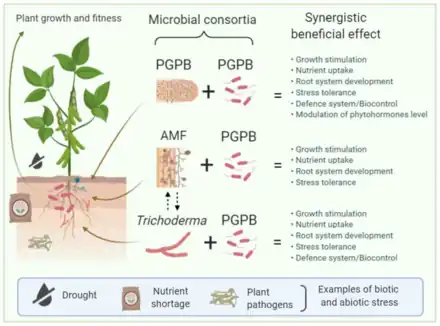
Although various studies have shown that single microorganisms can exert beneficial effects on plants, it is increasingly evident that when a microbial consortium — two or more interacting microorganisms — is involved, additive or synergistic results can be expected. This occurs, in part, due to the fact that multiple species can perform a variety of tasks in an ecosystem like the rhizosphere. Beneficial mechanisms of plant growth stimulation include enhanced nutrient availability, phytohormone modulation, biocontrol, biotic and abiotic stress tolerance) exerted by different microbial players within the rhizosphere, such as plant-growth-promoting bacteria (PGPB) and fungi such as Trichoderma and mycorrhizae.[47]
The diagram on the right illustrates that rhizosphere microorganisms like plant-growth-promoting bacteria (PGPB), arbuscular mycorrhizal fungi (AMF), and fungi from the genus Trichoderma spp. can establish beneficial interactions with plants, promoting plant growth and development, increasing the plant defense system against pathogens, promoting nutrient uptake, and enhancing tolerance to different environmental stresses. Rhizosphere microorganisms can influence one another, and the resulting consortia of PGPB + PGPB (e.g., a nitrogen-fixing bacterium such as Rhizobium spp. and Pseudomonas fluorescens), AMF + PGPB, and Trichoderma + PGPB may have synergetic effects on plant growth and fitness, providing the plant with enhanced benefits to overcome biotic and abiotic stress. Dashed arrows indicate beneficial interactions between AMF and Trichoderma.[47]
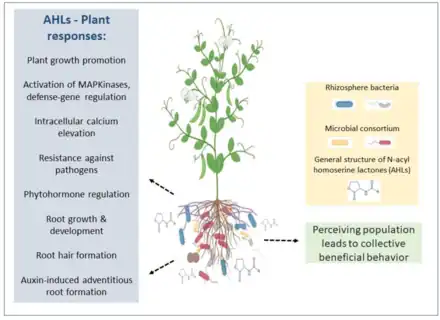
Communication
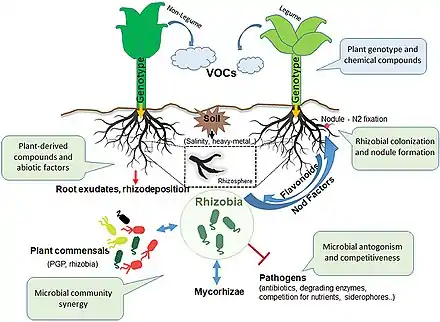
From bacterial quorum sensing to the signals of bees, communication is often the basis of biotic interactions. Frequently, more than two organisms can take part in the communication, resulting in a complex network of crosstalking. Recent advances in plant-microbe interactions research have shown that communication, both inter-kingdom and intra-kingdom, is shaped by a broad spectrum of factors. In this context, the rhizosphere (i.e., the soil close to the root surface) provides a specific microhabitat where complex interactions occur. The complex environment that makes up the rhizosphere can select for certain microbial populations, which are adapted to this unique niche. Among them, rhizobia have emerged as an important component of the rhizospheric microbiome. Rhizospheric crosstalk is found in rhizobium-legume interactions. This symbiosis is a complex process that involves signalling that can be shaped by plant rhizospheric exudates and microbiome composition. The relationship established by rhizobia with other rhizospheric organisms, together with the influence of the environmental factors, results in their beneficial role on host plant health.[48]
Prokaryotes and eukaryotes have interacted for millions of years, evolving and refining over time their communication systems. As proposed by Hauser in 1996,[10] biological signals and the exchange of information are part of the definition of communication, while the signals themselves are considered as "every structure able to shape the behavior of the organisms".[49][50] Consequently, the signals can evolve and persist thanks to the interaction between signals producers and receivers. Then, cooperation and fitness improvement are the basis of biological communication.[51][48]
In a particular environment, individuals can communicate and interact with multiple partners, and the nature of interaction can determine variable costs and benefits to the partner, as a biological market.[52] One of the most fascinating environments where complex biological interactions occur is the rhizosphere. Indeed, a large number of signals can be exchanged involving the plant itself, insects, fungi and microbes. This all take place in a high-density environmental niche. Usually, communication is the result of chemical responses of cells to signatory molecules coming from other cells. These signals affect both the metabolism and transcription of genes activating several regulatory mechanisms.[48]
Frequently in the rhizosphere, more than two organisms (and species) can take part in the communication, resulting in a complex network of interactions and cross-talks which influence the fitness of all participating partners. Thus, this environment can be considered a hot spot for numerous inter-kingdom signals exchange, which involves plant-associated microbial communities (rhizobiome). The microbial community's composition is mainly shaped and recruited by hundreds of metabolites released in the soil by plant roots, which normally facilitate the interactions with the biotic and abiotic environment. Often the plant can modulate their diversity based on the benefits in terms of growth and health, such as with plant growth-promoting rhizobacteria.[53] Nevertheless, a large number of nutrients issued by the plant can be of interest for the pathogenic organisms, which can take advantage of plant products for their survival in the rhizosphere.[54][48]
It stands to reason that the plants play a fundamental role in the rhizosphere scene.[55] Indeed, because of the chemical signals conveyed by nutrient-rich exudates released by the plant roots, a large variety of microbes can first colonize the rhizosphere and then gradually penetrate the root and the overall plant tissue (endophytes).[56] Otherwise, they can colonize the host plant establishing a lasting and beneficial symbiotic relationship.[57] To date, numerous investigations on root exudates composition have been performed.[53][58][59][48]
The most known plant-microbe dialogue on the rhizosphere scene, which determine direct and indirect advantages to the partners, was properly addressed as early as 1904 when Hiltner described the symbiotic interaction among legumes and rhizobia.[10] This symbiosis is a highly specific process in which the genetic and chemical communication signals are strictly plant-bacterium specific. In this mutualistic interaction, rhizobia positively influence the host's growth thanks to the nitrogen fixation process and at the same time can benefit from the nutrients provided by the plant.[48]
This symbiosis has been extensively studied in recent decades, and many studies on the communication and the signaling between the two partners at different steps of the symbiosis (from root infection to nodules development) have been elucidated.[60][61] However, the knowledge about the earlier steps of rhizosphere colonization, namely the opening line at the root surface, remains poorly characterized. Nonetheless, increasing data have shown the importance of intraspecies and multispecies communications among rhizospheric biotic components for the improvement of rhizobia–legumes interaction. In addition, it has been shown that rhizobia are part of the rhizosphere of a wide variety of non-legume plants where they can play a role as plant growth promoting components, recovering a central role in plant core microbiome.[62][48]
Methods
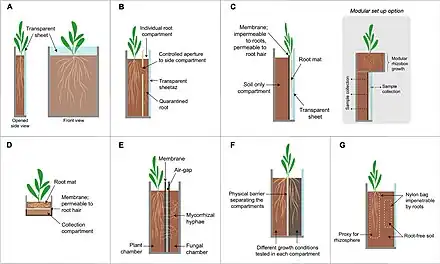
Following are some methods commonly used or of interest in rhizosphere research. Many of these methods include both field testing of the root systems and in lab testing using simulated environments to perform experiments, such as pH determination.[63]
- High-throughput screening
- High-Throughput Sequencing: 16S rRNA Amplicon, Metagenomics, Metatranscriptomics
- Culture Depend Approaches
- Root Imaging
- Isotopic labeling
- Enzyme Assays
- Mini rhizotron camera
- Various methods used to determine water movement in the rhizosphere e.g. microelectrodes and agar techniques for pH and microsampling of rhizosphere materials[63]
- Pyrolysis–field ionization mass spectrometry allows for spectrometry of agricultural fields to find fulvic and humic acids and the extraction residues (humins) in certain studies and expanded to general organic compounds in other recent work.[64][21]
See also
References
- Yee, Mon Oo; Kim, Peter; Li, Yifan; Singh, Anup K.; Northen, Trent R.; Chakraborty, Romy (26 March 2021). "Specialized Plant Growth Chamber Designs to Study Complex Rhizosphere Interactions". Frontiers in Microbiology. Frontiers Media SA. 12: 625752. doi:10.3389/fmicb.2021.625752. ISSN 1664-302X. PMC 8032546. PMID 33841353.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - "Microbial Health of the Rhizosphere". Archived from the original on March 12, 2007. Retrieved 5 May 2006.
- Hütsch, Birgit W.; Augustin, Jürgen; Merbach, Wolfgang (2002). "Plant rhizodeposition – an important source for carbon turnover in soils". Journal of Plant Nutrition and Soil Science. 165 (4): 397–407. doi:10.1002/1522-2624(200208)165:4<397::AID-JPLN397>3.0.CO;2-C – via Research Gate.
- Walker, T. S.; Bais, H. P.; Grotewold, E.; Vivanco, J. M. (2003). "Root exudation and rhizosphere biology". Plant Physiology. 132 (1): 44–51. doi:10.1104/pp.102.019661. PMC 1540314. PMID 12746510.
- Elaine R. Ingham. "The Soil Food Web". USDA-NRCS. Retrieved 3 July 2006.
- Grayston, Susan J.; Wang, Shenquiang; Campbell, Colin D.; Edwards, Anthony C. (March 1998). "Selective influence of plant species on microbial diversity in the rhizosphere". Soil Biology and Biochemistry. 30 (3): 369–378. doi:10.1016/S0038-0717(97)00124-7.
- Estermann, Eva F.; McLaren, A. D. (1961). "Contribution of rhizoplane organisms to the total capacity of plants to utilize organic nutrients". Plant and Soil. 15 (3): 243–260. doi:10.1007/BF01400458. S2CID 35099987.
- Mc Near Jr., D. H. (2013). "The Rhizosphere - roots, soil and everything in between". Nature Education. 4 (3): 1.
- Hiltner, L. (1904) "Ueber neuere Erfahrungen und Probleme auf dem Gebiete derBodenbakteriologie und unter besonderer BerUcksichtigung der Grundungung und Brache. Arb Deut Landw Gesell, 98: 57-78.
- Hartmann, Anton; Rothballer, Michael; Schmid, Michael (2008). "Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research". Plant and Soil. 312 (1–2): 7–14. doi:10.1007/s11104-007-9514-z. S2CID 4419735.
- McNear Jr., D. H. (2013) "The Rhizosphere - Roots, Soil and Everything In Between". Nature Education Knowledge, 4(3): 1.
- Canarini A, Kaiser C, Merchant A, Richter A and Wanek W (2019) Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Frontiers in Plant Science 10:157. doi: 10.3389/fpls.2019.00157
- Jones, David L. (August 1998). "Organic acids in the rhizosphere – a critical review". Plant and Soil. 205 (1): 25–44. doi:10.1023/A:1004356007312. S2CID 26813067.
- Hinsinger, Philippe (December 2001). "Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review". Plant and Soil. 237 (2): 173–195. doi:10.1023/A:1013351617532. S2CID 8562338.
- Walker, Travis S.; Bais, Harsh Pal; Grotewold, Erich; Vivanco, Jorge M. (May 2003). "Root Exudation and Rhizosphere Biology". Plant Physiol. 132 (1): 44–51. doi:10.1104/pp.102.019661. PMC 1540314. PMID 12746510.
- Lambers, Hans; Mougel, Christophe; Jaillard, Benoît; Hinsinger, Philippe (August 2009). "Plant-microbe-soil interactions in the rhizosphere: an evolutionary perspective". Plant Soil. 321 (1–2): 83–115. doi:10.1007/s11104-009-0042-x. S2CID 6840457.
- Hinsinger, Philippe; Gobran, George R.; Gregory, Peter J.; Wenzel, Walter W. (November 2005). "Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes". New Phytologist. 168 (2): 293–303. doi:10.1111/j.1469-8137.2005.01512.x. ISSN 1469-8137. PMID 16219069.
- Czarnes, S.; Hallett, P. D.; Bengough, A. G.; Young, I. M. (2000). "Root- and microbial-derived mucilages affect soil structure and water transport". European Journal of Soil Science. 51 (3): 435. doi:10.1046/j.1365-2389.2000.00327.x. S2CID 96925936.
- Bérard, Annette; Clavel, Thierry; Le Bourvellec, Carine; Davoine, Aurélien; Le Gall, Samuel; Doussan, Claude; Bureau, Sylvie (2020). "Exopolysaccharides in the rhizosphere: A comparative study of extraction methods. Application to their quantification in Mediterranean soils". Soil Biology and Biochemistry. 149: 107961. doi:10.1016/j.soilbio.2020.107961.
- Le Gall, Samuel; Bérard, Annette; Page, David; Lanoe, Lucas; Bertin, Nadia; Doussan, Claude (2021). "Increased exopolysaccharide production and microbial activity affect soil water retention and field performance of tomato under water deficit". Rhizosphere. 19: 100408. doi:10.1016/j.rhisph.2021.100408. ISSN 2452-2198.
- Schlichting, André; Leinweber, Peter (2009). "New evidence for the molecular–chemical diversity of potato plant rhizodeposits obtained by pyrolysis–field Ionisation mass spectrometry". Phytochemical Analysis. 20 (1): 1–13. doi:10.1002/pca.1080. ISSN 1099-1565. PMID 18618895.
- Leslie A. Weston & Ulrike Mathesius (2013). "Flavonoids: Their Structure, Biosynthesis and Role in the Rhizosphere, Including Allelopathy". Journal of Chemical Ecology. 39 (2): 283–297. doi:10.1007/s10886-013-0248-5. PMID 23397456. S2CID 14121251.
- Bertin, Cecile; Yang, Xiaohan; Weston, Leslie A. (2003). "The role of root exudates and allelochemicals in the rhizosphere". Plant and Soil. Springer Science and Business Media LLC. 256 (1): 67–83. doi:10.1023/a:1026290508166. ISSN 0032-079X. S2CID 9066759.
- Glass, Anthony D. M. (1976). "The allelopathic potential of phenolic acids associated with the rhizosphere of Pteridium aquilinum". Canadian Journal of Botany. 54 (21): 2440–2444. doi:10.1139/b76-259.
- Bais, Harsh P.; Weir, Tiffany L.; Perry, Laura G.; Gilroy, Simon; Vivanco, Jorge M. (2006). "The Role of Root Exudates in Rhizosphere Interactions with Plants and Other Organisms". Annual Review of Plant Biology. 57 (1): 233–266. doi:10.1146/annurev.arplant.57.032905.105159. PMID 16669762.
- van Loon, L. C.; Bakker, P. A. H. M.; Pieterse, C. M. J. (28 November 2003). "Systemic Resistance Induced by Rhizosphere Bacteria". Annual Review of Phytopathology. 36 (1): 453–483. doi:10.1146/annurev.phyto.36.1.453. hdl:1874/7701. PMID 15012509.
- Stinson, Kristina A; Campbell, Stuart A; Powell, Jeff R; Wolfe, Benjamin E; Callaway, Ragan M; Thelen, Giles C; Hallett, Steven G; Prati, Daniel; Klironomos, John N (2006-04-25). Loreau, Michel (ed.). "Invasive Plant Suppresses the Growth of Native Tree Seedlings by Disrupting Belowground Mutualisms". PLOS Biology. Public Library of Science (PLoS). 4 (5): e140. doi:10.1371/journal.pbio.0040140. ISSN 1545-7885. PMC 1440938. PMID 16623597.
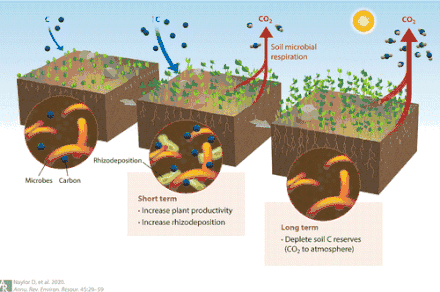
- Naylor, Dan; Sadler, Natalie; Bhattacharjee, Arunima; Graham, Emily B.; Anderton, Christopher R.; McClure, Ryan; Lipton, Mary; Hofmockel, Kirsten S.; Jansson, Janet K. (2020-10-17). "Soil Microbiomes Under Climate Change and Implications for Carbon Cycling". Annual Review of Environment and Resources. Annual Reviews. 45 (1): 29–59. doi:10.1146/annurev-environ-012320-082720. ISSN 1543-5938. S2CID 219905513.
 Modified material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Jansson, Janet K.; Taş, Neslihan (2014-05-12). "The microbial ecology of permafrost". Nature Reviews Microbiology. Springer Science and Business Media LLC. 12 (6): 414–425. doi:10.1038/nrmicro3262. ISSN 1740-1526. PMID 24814065. S2CID 42795586.
- Mackelprang, Rachel; Saleska, Scott R.; Jacobsen, Carsten Suhr; Jansson, Janet K.; Taş, Neslihan (2016-06-29). "Permafrost Meta-Omics and Climate Change". Annual Review of Earth and Planetary Sciences. Annual Reviews. 44 (1): 439–462. doi:10.1146/annurev-earth-060614-105126. ISSN 0084-6597. S2CID 131260721.
- Moore, John C.; McCann, Kevin; Setälä, Heikki; De Ruiter, Peter C. (2003). "Top-Down is Bottom-Up: Does Predation in the Rhizosphere Regulate Aboveground Dynamics?". Ecology. 84 (4): 846. doi:10.1890/0012-9658(2003)084[0846:TIBDPI]2.0.CO;2.
- Ingham, Russell E.; Trofymow, J. A.; Ingham, Elaine R.; Coleman, David C. (1985). "Interactions of Bacteria, Fungi, and their Nematode Grazers: Effects on Nutrient Cycling and Plant Growth". Ecological Monographs. 55 (1): 119–140. doi:10.2307/1942528. JSTOR 1942528.
- Jousset, Alexandre; Rochat, Laurène; Scheu, Stefan; Bonkowski, Michael; Keel, Christoph (August 2010). "Predator-Prey Chemical Warfare Determines the Expression of Biocontrol Genes by Rhizosphere-Associated Pseudomonas fluorescens". Appl. Environ. Microbiol. 76 (15): 5263–5268. Bibcode:2010ApEnM..76.5263J. doi:10.1128/AEM.02941-09. PMC 2916451. PMID 20525866.
- Bais, Harsh Pal; Park, Sang-Wook; Weir, Tiffany L; Callaway, Ragan M; Vivanco, Jorge M (2004). "How plants communicate using the underground information superhighway". Trends in Plant Science. 9 (1): 26–32. doi:10.1016/j.tplants.2003.11.008. PMID 14729216.
- Raynaud, Xavier; Jaillard, Benoît; Leadley, Paul W. (2008). "Plants May Alter Competition by Modifying Nutrient Bioavailability in Rhizosphere: A Modeling Approach" (PDF). The American Naturalist. 171 (1): 44–58. doi:10.1086/523951. ISSN 0003-0147. PMID 18171150. S2CID 23413577.
- Kaye, Jason P.; Hart, Stephen C. (1997). "Competition for nitrogen between plants and soil microorganisms". Trends in Ecology & Evolution. 12 (4): 139–43. doi:10.1016/S0169-5347(97)01001-X. PMID 21238010.
- Owen, A.G; Jones, D.L (2001). "Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition". Soil Biology and Biochemistry. 33 (4–5): 651–657. doi:10.1016/s0038-0717(00)00209-1.
- Bodegom, Peter van; Stams, Fons; Mollema, Liesbeth; Boeke, Sara; Leffelaar, Peter (August 2001). "Methane Oxidation and the Competition for Oxygen in the Rice Rhizosphere". Appl. Environ. Microbiol. 67 (8): 3586–3597. Bibcode:2001ApEnM..67.3586V. doi:10.1128/AEM.67.8.3586-3597.2001. PMC 93059. PMID 11472935.
- McCann, Kevin; Hastings, Alan; Huxel, Gary R. (22 October 1998). "Weak trophic interactions and the balance of nature". Nature. 395 (6704): 794–798. Bibcode:1998Natur.395..794M. doi:10.1038/27427. S2CID 4420271.
- Besserer, Arnaud; Puech-Pagès, Virginie; Kiefer, Patrick; Gomez-Roldan, Victoria; Jauneau, Alain; Roy, Sébastien; Portais, Jean-Charles; Roux, Christophe; Bécard, Guillaume; Séjalon-Delmas, Nathalie (2006-06-27). Chory, Joanne (ed.). "Strigolactones Stimulate Arbuscular Mycorrhizal Fungi by Activating Mitochondria". PLOS Biology. Public Library of Science (PLoS). 4 (7): e226. doi:10.1371/journal.pbio.0040226. ISSN 1545-7885. PMC 1481526. PMID 16787107.
- Andreas Brachmann, Martin Parniske (2006). "The Most Widespread Symbiosis on Earth". PLOS Biology. 4 (7): e239. doi:10.1371/journal.pbio.0040239. PMC 1489982. PMID 16822096.
- Tian, C. F.; Garnerone, A.-M.; Mathieu-Demaziere, C.; Masson-Boivin, C.; Batut, J. (2012). "Plant-activated bacterial receptor adenylate cyclases modulate epidermal infection in the Sinorhizobium meliloti-Medicago symbiosis". Proceedings of the National Academy of Sciences. 109 (17): 6751–6756. Bibcode:2012PNAS..109.6751T. doi:10.1073/pnas.1120260109. PMC 3340038. PMID 22493242.
- Sims GK, Dunigan EP; Dunigan (1984). "Diurnal and seasonal variations in nitrogenase activity (C2H2 reduction) of rice roots". Soil Biology and Biochemistry. 16 (1): 15–18. doi:10.1016/0038-0717(84)90118-4.
- Weller, D. M. (1988). "Biological Control of Soilborne Plant Pathogens in the Rhizosphere with Bacteria". Annual Review of Phytopathology. 26 (1): 379–407. doi:10.1146/annurev.py.26.090188.002115.
- Bianciotto, V.; Minerdi, D.; Perotto, S.; Bonfante, P. (1996). "Cellular interactions between arbuscular mycorrhizal fungi and rhizosphere bacteria". Protoplasma. 193 (1–4): 123–131. doi:10.1007/BF01276640. S2CID 39961232.
- Howell, C. R. (2003). "Mechanisms Employed by Trichoderma Species in the Biological Control of Plant Diseases: The History and Evolution of Current Concepts". Plant Disease. 87 (1): 4–10. doi:10.1094/PDIS.2003.87.1.4. PMID 30812698.
- Santoyo, Gustavo; Guzmán-Guzmán, Paulina; Parra-Cota, Fannie Isela; Santos-Villalobos, Sergio de los; Orozco-Mosqueda, Ma. del Carmen; Glick, Bernard R. (2021). "Plant Growth Stimulation by Microbial Consortia". Agronomy. 11 (2): 219. doi:10.3390/agronomy11020219.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Checcucci, Alice; Marchetti, Marta (2020). "The Rhizosphere Talk Show: The Rhizobia on Stage". Frontiers in Agronomy. 2. doi:10.3389/fagro.2020.591494.
 Material was copied from this source, which is available under a .
Material was copied from this source, which is available under a . - Maynard-Smith, John; Harper, David (6 November 2003). Animal Signals. New York. ISBN 978-0-19-852685-8. OCLC 54460090.
{{cite book}}: CS1 maint: location missing publisher (link) - Scott-Phillips, T. C. (2008). "Defining biological communication". Journal of Evolutionary Biology. 21 (2): 387–395. doi:10.1111/j.1420-9101.2007.01497.x. PMID 18205776. S2CID 5014169.
- Zahavi, A. (2008). "The handicap principle and signalling in collaborative systems". In: Sociobiology of Communication, eds P. d'Ettorre and D. P. Hughes (Oxford University Press), 1–10. chapter 1
- Werner, Gijsbert D. A.; Cornwell, William K.; Cornelissen, Johannes H. C.; Kiers, E. Toby (2015). "Evolutionary signals of symbiotic persistence in the legume–rhizobia mutualism". Proceedings of the National Academy of Sciences. 112 (33): 10262–10269. Bibcode:2015PNAS..11210262W. doi:10.1073/pnas.1424030112. PMC 4547229. PMID 26041807.
- Hartmann, Anton; Schmid, Michael; Tuinen, Diederik van; Berg, Gabriele (2009). "Plant-driven selection of microbes". Plant and Soil. 321 (1–2): 235–257. doi:10.1007/s11104-008-9814-y. S2CID 17890501.
- Rasmann, Sergio; Turlings, Ted CJ (2016). "Root signals that mediate mutualistic interactions in the rhizosphere". Current Opinion in Plant Biology. 32: 62–68. doi:10.1016/j.pbi.2016.06.017. PMID 27393937.
- Bending, G.D. (2017). "The Rhizopshere and Its Microorganisms". Encyclopedia of Applied Plant Sciences. pp. 347–351. doi:10.1016/B978-0-12-394807-6.00165-9. ISBN 9780123948083.
- Hardoim, Pablo R.; Van Overbeek, Leo S.; Elsas, Jan Dirk van (2008). "Properties of bacterial endophytes and their proposed role in plant growth". Trends in Microbiology. 16 (10): 463–471. doi:10.1016/j.tim.2008.07.008. PMID 18789693.
- Chi, Feng; Shen, Shi-Hua; Cheng, Hai-Ping; Jing, Yu-Xiang; Yanni, Youssef G.; Dazzo, Frank B. (2005). "Ascending Migration of Endophytic Rhizobia, from Roots to Leaves, inside Rice Plants and Assessment of Benefits to Rice Growth Physiology". Applied and Environmental Microbiology. 71 (11): 7271–7278. Bibcode:2005ApEnM..71.7271C. doi:10.1128/AEM.71.11.7271-7278.2005. PMC 1287620. PMID 16269768.
- Bulgarelli, Davide; Schlaeppi, Klaus; Spaepen, Stijn; Van Themaat, Emiel Ver Loren; Schulze-Lefert, Paul (2013). "Structure and Functions of the Bacterial Microbiota of Plants". Annual Review of Plant Biology. 64: 807–838. doi:10.1146/annurev-arplant-050312-120106. PMID 23373698.
- Venturi, Vittorio; Keel, Christoph (2016). "Signaling in the Rhizosphere". Trends in Plant Science. 21 (3): 187–198. doi:10.1016/j.tplants.2016.01.005. PMID 26832945.
- Oldroyd, Giles E.D.; Murray, Jeremy D.; Poole, Philip S.; Downie, J. Allan (2011). "The Rules of Engagement in the Legume-Rhizobial Symbiosis". Annual Review of Genetics. 45: 119–144. doi:10.1146/annurev-genet-110410-132549. PMID 21838550.
- Oldroyd, Giles E. D. (2013). "Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants". Nature Reviews Microbiology. 11 (4): 252–263. doi:10.1038/nrmicro2990. PMID 23493145. S2CID 7849557.
- Yeoh, Yun Kit; Paungfoo-Lonhienne, Chanyarat; Dennis, Paul G.; Robinson, Nicole; Ragan, Mark A.; Schmidt, Susanne; Hugenholtz, Philip (2016). "The core root microbiome of sugarcanes cultivated under varying nitrogen fertilizer application". Environmental Microbiology. 18 (5): 1338–1351. doi:10.1111/1462-2920.12925. PMID 26032777.
- Gregory, P.J.; Hinsinger, P. (1999). "New approaches to studying chemical and physical changes in the rhizosphere: an overview". Plant and Soil. 211: 1–9. doi:10.1023/A:1004547401951. S2CID 24345065.
- Schlten, Hans-Rolf; Leinweber, Peter (April 1993). "Pyrolysis-field ionization mass spectrometry of agricultural soils and humic substances: Effect of cropping systems and influence of the mineral matrix". Plant and Soil. 151 (1): 77–90. doi:10.1007/BF00010788. ISSN 0032-079X. S2CID 23547420.
- Giri, B.; Giang, P. H.; Kumari, R.; Prasad, R.; Varma, A. (2005). "Microbial Diversity in Soils". Microorganisms in Soils: Roles in Genesis and Functions. Soil Biology. Vol. 3. pp. 19–55. doi:10.1007/3-540-26609-7_2. ISBN 978-3-540-22220-0.
Further reading
- "The Soil Habitat". University of Western Australia. Archived from the original on 20 August 2006. Retrieved 3 July 2006.
- Andrew Wylie (2006). "Digging in the Dirt: Is the Study of the Rhizosphere Ripe for a Systems Biology Approach?". Science Creative Quarterly. Retrieved 24 April 2017.
- McNear Jr., D. H. (2013). "The Rhizosphere – Roots, Soil and Everything In Between". Nature Education Knowledge. 4 (3): 1. Retrieved 24 April 2017.