SOGA2
SOGA2, also known as Suppressor of glucose autophagy associated 2 or CCDC165, is a protein that in humans is encoded by the SOGA2 gene.[5][6] SOGA2 has two human paralogs, SOGA1 and SOGA3.[7][8] In humans, the gene coding sequence is 151,349 base pairs long, with an mRNA of 6092 base pairs, and a protein sequence of 1586 amino acids. The SOGA2 gene is conserved in gorilla, baboon, galago, rat, mouse, cat, and more. There is distant conservation seen in organisms such as zebra finches and anoles.[9] SOGA2 is ubiquitously expressed in humans, with especially high expression in brain (especially the cerebellum and hippocampus), colon, pituitary gland, small intestine, spinal cord, testis and fetal brain.[10]
| MTCL1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MTCL1, CCDC165, KIAA0802, SOGA2, microtubule crosslinking factor 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 615766 MGI: 1915867 HomoloGene: 41017 GeneCards: MTCL1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Gene
Locus
The SOGA2 gene is located from 8717369 - 8832775 on the short arm of chromosome 18 (18p11.22).[11]
Homology and Evolution
Paralogs
There are two main paralogs to SOGA2: human protein SOGA1 and human protein SOGA3.[9] SOGA1 has been shown to be involved in suppression of glucose by autophagy.[12] The rate at which orthologs diverge from SOGA2 human(measured by % identity) places the approximate duplication event of SOGA1 from SOGA2 at ~254.1 MYA and the duplication event of SOGA3 from SOGA2 ~329.1 MYA.
| protein name | accession number | sequence length (aa) | sequence identity to human protein | notes |
| SOGA3 | NP_001012279.1 | 947 | 58% | conserved in ~500 N-terminal aa |
| SOGA1 isoform 2 | NP_954650.2 | 1016 aa | 65% | conserved in first ~900 aa |
| SOGA1 isoform 1 | NP_542194.2 | 1661 | 41% | conserved across the length of sequence except ~950-1150 |
Orthologs
Many orthologs have been identified in Eukaryotes.[9]
| common name | protein name | divergence from human lineage (MYA) | accession number | sequence length (aa) | sequence identity to human protein | protein domain differences |
|---|---|---|---|---|---|---|
| gorilla | protein SOGA2 | 8.8 | XP_004059220.1 | 1586 | 99% | |
| baboon | protein SOGA2 | 29 | XP_003914218 | 1587 | 98% | |
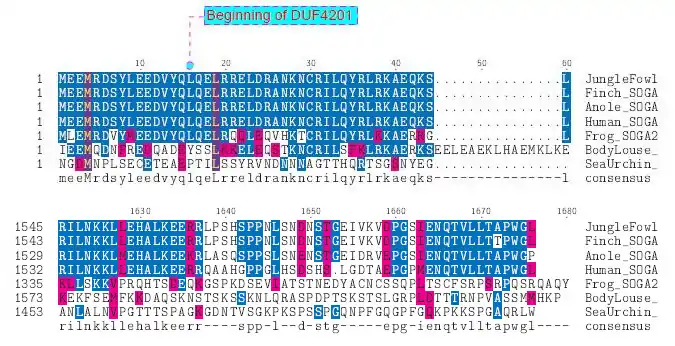
| galago | protein SOGA3 | 74 | XP_003801047.1 | 1583 | 88% | DUF4201 not present |
| rat | CCDC165 | 92.3 | XP_237548.6 | 2060 | 81% | DUF4201 not present |
| mouse | SOGA2 | 92.3 | NP_001107570.1 | 1893 | 80% | |
| house cat | protein SOGA2 | 94.2 | XP_003995077.1 | 1700 | 84% | DUF4201 not present |
| cow | CCDC166 | 94.2 | XP_581047.5 | 1525 | 74% | DUF4201 not present |
| African Elephant | CCDC167-like | 98.7 | XP_003406836.1 | 1544 | 73% | |
| zebra Finch | protein SOGA2 | 296 | XP_002193121.1 | 1598 | 69% | DUF4201 not present |
| Red JungleFowl | CCDC165 | 296 | XP_423729.3 | 1600 | 70% | DUF4201 not present |
| Carolina anole | uncharacterized protein KIAA0802-like | 296 | XP_003225723.1 | 1839 | 67% | DUF4201 not present |
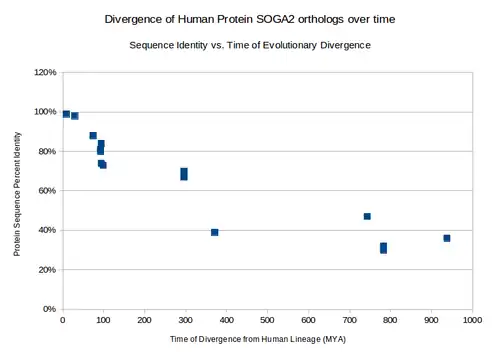
Distant Homologs
| common name | protein name | divergence from human lineage (MYA) | accession number | sequence length (aa) | sequence identity to human protein | protein domain differences |
|---|---|---|---|---|---|---|
| Tropical Clawed Frog | uncharacterized protein C20orf117-like | 371.2 | XP_002942331.1 | 1584 | 39% | |
| purple sea urchin | uncharacterized protein LOC578090 | 742.9 | XP_783370.2 | 1587 | 47% | DUF4201 not present |
| body louse | Centromeric protein E, putative | 782.7 | XP_002429877.1 | 2086 | 30% | no shared domains |
| southern house mosquito | conserved hypothetical protein | 782.7 | XP_001843754.1 | 1878 | 32% | no shared domains |
| porkworm | surface antigen repeat family protein | 937.5 | XP_003380263.1 | 2030 | 36% | no shared domains |
Homologous Domains
SOGA2 is conserved farthest back in its N-terminal region, where it contains its three domains of unknown function.[13]
Protein
Protein internal composition
SOGA2 is rich in glycine (ratio r of SOGA2 composition to average human protein is 1.723), glutamate (r = 1.647), and arginine (r = 1.357). It also has a lower than usual composition of tyrosine (r = 0.3406), isoleucine (r = 0.4430), phenylalanine (r = 0.5808), and valine (r = 0.6161).[14][15]
Primary structure and isoforms
SOGA2 has 4 isoforms: Q9Y4B5-1, Q9Y4B5-2, Q9Y4B5-3, Q9Y4B5-4.[16]
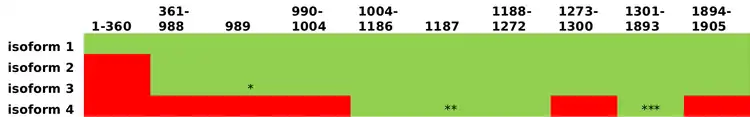
Domains and motifs
SOGA2 contains Domain of Unknown Function 4201 (DUF4201) from aa 16-235. This domain is specific to the Coiled Coil Domain Containing family of proteins in eukaryotes.[17] It also contains two copies of Domain of Unknown Function 3166 (DUF3166): one from aa 140-235 and one from aa 269-364.[11]
Post-translational modifications
SOGA2 is expected to undergo a number of post-translational modifications. Modifications of human SOGA2 that are shared by orthologs include:
- Sumoylation at amino acids 87, 152, 235, 392, and 1379.[18]
- Sulfination at tyrosines 14 and 1249.[19]
- Phosphorylation at a number of sites, highlighted in the following graphic:

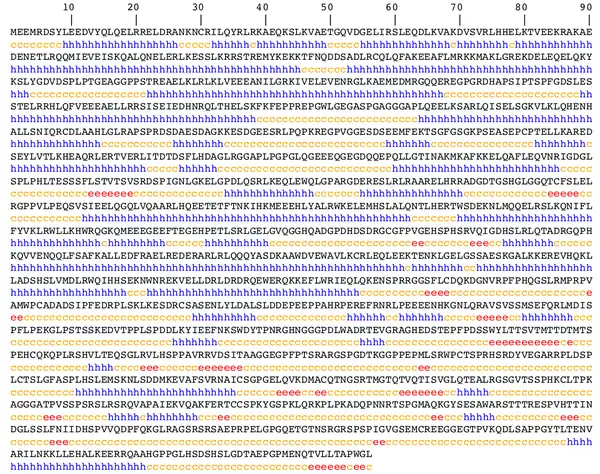
Secondary structure
The consensus of the prediction software PELE,[21] GOR4,[22] and SOSUICoil is that the secondary structure of SOGA2 is dominated by alpha helices with interspersed regions of random coil. GOR4 indicated that SOGA2 is dominated by alpha-helices; it predicted a mere 5.61% of residues in an extended strand (parallel or antiparallel Beta-sheet) conformation, as opposed to 47.79% alpha helix and 46.6% random coils.
Tertiary structure
SOGA2 shares sequence features in its highly conserved N-terminal region. This homology allows prediction of its tertiary structure on the basis of homology to published 3d structures via Phyre2[24] and NCBI structure.[25]
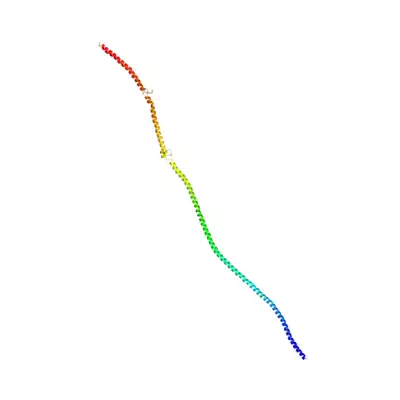 SOGA2's 3d structure predicted by Phyre2.[24] Structure is based on the crystal structure of tropomyosin at 7 angstrom resolution, with 12% identity. 283 residues match, in the CCDC containing N-terminal region. |
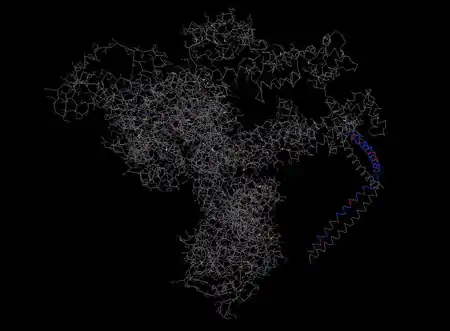 1I84 S, Heavy Meromyosin Subfragment Of Chicken Gizzard Smooth Muscle Myosin With Regulatory Light Chain In The Dephosphorylated State 3d structure. Highlighted region is conserved in SOGA2.[25] |
Gene expression
Promoter
The promoter for human SOGA2 is below.

Gene expression data
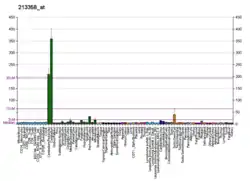
The EST profile shows that, in humans, SOGA2 is highly expressed in many sites throughout the body, including bone, brain, ear, eye, and many others.[26] There are a large number of transcripts in liver cancer samples. Human microarray data show that SOGA2 is moderately expressed, with especially high expression in brain (especially the cerebellum and hippocampus), colon, pituitary gland, small intestine, spinal cord, testis and fetal brain.[10] Brain-tissue-specific microarray data show that SOGA2 has high expression throughout the posterior lobe of the cerebellar hemispheres and posterial lobe of the vermis in the mouse brain. There is low expression in most other areas of the brain.[27]
Transcript variants
In humans, the SOGA2 gene produces 17 different transcripts, 8 of which form a protein product (one undergoes nonsense mediated decay). The main transcript in humans is transcript ID ENST00000359865, or SOGA2-001.[28]
Function
Possible transcription factors
Possible transcription factors for human SOGA2 include:[29]
- Modulator recognition factor 2
- cAMP-responsive element binding protein 1
- alternative splicing variant of FOXP1
- MDS1/EVI1-like gene 1
- Ikaros 2, possible regulator of lymphocyte differentiation
Interactions
Protein complex co-immunoprecipitation (Co-IP) experiments revealed interacting proteins such as cell death regulators, ATP-binding cassette (ABC) transporters and protein kinase A binding proteins.[30]
The 540 interacting proteins include ABCF1, ACTB, ACTL6A, BCLAF1, BCLAF1, CHEK1, and MAGEE2.[30]
K-nearest neighbor analysis by wolf pSort indicates that in humans, SOGA2 is focused mainly in the nucleus, cytoplasm, and the cytonuclear space. There is a small chance that it is localizes to the golgi.[31]
A number of protein interactants were also identified via the STRING database, including MARK2, MARK4, and PPP2R2B.
Clinical significance
SOGA2 has no currently known disease associations or mutations.
References
- GRCh38: Ensembl release 89: ENSG00000168502 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000052105 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Nagase T; Ishikawa K; Suyama M; Kikuno R; Miyajima N; Tanaka A; Kotani H; Nomura N; Ohara O (April 1999). "Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Res. 5 (5): 277–86. doi:10.1093/dnares/5.5.277. PMID 9872452.
- "Entrez Gene: SOGA2".
- "SOGA1". NCBI. Retrieved April 27, 2013.
- "SOGA3". NCBI. Retrieved April 27, 2013.
- "BLAST". NCBI BLAST. Retrieved April 27, 2013.
- "GEO Profile 10132039". NCBI GEO. Retrieved April 27, 2013.
- "NCBI". National Center for Biotechnology Information. Retrieved 12 May 2013.
- Cowherd RB, Cowerd RB, Asmar MM, et al. (October 2010). "Adiponectin lowers glucose production by increasing SOGA". Am. J. Pathol. 177 (4): 1936–45. doi:10.2353/ajpath.2010.100363. PMC 2947288. PMID 20813965.
- "CLUSTALW". SDSC Biology Workbench. Retrieved April 27, 2013.
- "CLC Sequence Viewer". Retrieved 12 May 2011.
- Nagase T; Ishikawa K; Suyama M; Kikuno R; Miyajima N; Tanaka A; Kotani H; Nomura N; Ohara O (Jan 2011). "Computational analysis of amino acid composition in human proteins". Bioinformatics Trends. 6 (1&2): 39–43.
- "GeneCards". Retrieved 9 May 2011.
- "NCBI Conserved Domains". National Center for Biotechnology Information. Retrieved 12 May 2013.
- "SumoPlot". ABGENT. Retrieved April 27, 2013.
- "Sulfinator". expasy. Retrieved April 27, 2013.
- Blom N; Gammeltoft S; Brunak S (December 1999). "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites". J. Mol. Biol. 294 (5): 1351–62. doi:10.1006/jmbi.1999.3310. PMID 10600390.
- "PELE". SDSC Biology Workbench. Retrieved 27 April 2013.
- "GOR4". npsa-pbil. Retrieved 27 April 2013.
- "SOSUICoil". bp.nuap.nagoya-u.ac.jp. Archived from the original on 2011-07-22. Retrieved 27 April 2013.
- Kelley LA; Sternberg MJ (2009). "Protein structure prediction on the Web: a case study using the Phyre server" (PDF). Nat Protoc. 4 (3): 363–71. doi:10.1038/nprot.2009.2. hdl:10044/1/18157. PMID 19247286. S2CID 12497300.
- "NCBI Structure". NCBI. Retrieved May 13, 2013.
- "Unigene". National Center for Biotechnology Information. Retrieved April 27, 2013.
- "Allen Brain Atlas, SOGA2 microarray experiments". Allen Brain Atlas. Retrieved April 27, 2013.
- "Ensemble: gene SOGA2". Ensembl. Retrieved April 27, 2013.
- "El Dorado". Genomatix. Retrieved May 8, 2013.
- "Molecular Interaction Database - MINT". Archived from the original on 6 May 2006. Retrieved 9 May 2011.
- Horton P, Park KJ, Obayashi T, et al. (July 2007). "WoLF PSORT: protein localization predictor". Nucleic Acids Res. 35 (Web Server issue): W585–7. doi:10.1093/nar/gkm259. PMC 1933216. PMID 17517783.
Further reading
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Brajenovic M, Joberty G, Küster B, et al. (2004). "Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network". J. Biol. Chem. 279 (13): 12804–11. doi:10.1074/jbc.M312171200. PMID 14676191.
- Ota T, Suzuki Y, Nishikawa T, et al. (2004). "Complete sequencing and characterization of 21,243 full-length human cDNAs". Nat. Genet. 36 (1): 40–5. doi:10.1038/ng1285. PMID 14702039.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Nusbaum C, Zody MC, Borowsky ML, et al. (2005). "DNA sequence and analysis of human chromosome 18". Nature. 437 (7058): 551–5. Bibcode:2005Natur.437..551N. doi:10.1038/nature03983. PMID 16177791.
- Nousiainen M, Silljé HH, Sauer G, et al. (2006). "Phosphoproteome analysis of the human mitotic spindle". Proc. Natl. Acad. Sci. U.S.A. 103 (14): 5391–6. Bibcode:2006PNAS..103.5391N. doi:10.1073/pnas.0507066103. PMC 1459365. PMID 16565220.
- Beausoleil SA, Villén J, Gerber SA, et al. (2006). "A probability-based approach for high-throughput protein phosphorylation analysis and site localization". Nat. Biotechnol. 24 (10): 1285–92. doi:10.1038/nbt1240. PMID 16964243. S2CID 14294292.
- Olsen JV, Blagoev B, Gnad F, et al. (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983. S2CID 7827573.