Staphylococcus aureus
Staphylococcus aureus is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen.[1] Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.
| Staphylococcus aureus | |
|---|---|
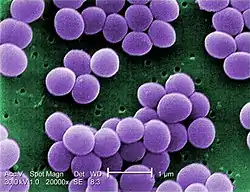 | |
| Scanning electron micrograph of S. aureus; false color added | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Bacillota |
| Class: | Bacilli |
| Order: | Bacillales |
| Family: | Staphylococcaceae |
| Genus: | Staphylococcus |
| Species: | S. aureus |
| Binomial name | |
| Staphylococcus aureus Rosenbach 1884 | |
| Staphylococcus aureus | |
|---|---|
| Other names | Staph aureus, S. aureus |
| Specialty | Infectious disease |
| Types | Methicillin-susceptible Staphylococcus aureus, Methicillin-resistant Staphylococcus aureus |
| Causes | Staphylococcus aureus bacteria |
| Differential diagnosis | other bacterial, viral and fungal infections, |
| Prevention | hand washing, cleaning surfaces |
| Medication | Antibiotics |
| Frequency | 20% to 30% of the human population often without symptoms |
An estimated 21% to 30% of the human population are long-term carriers of S. aureus,[2][3] which can be found as part of the normal skin microbiota, in the nostrils,[2][4] and as a normal inhabitant of the lower reproductive tract of females.[5][6] S. aureus can cause a range of illnesses, from minor skin infections, such as pimples,[7] impetigo, boils, cellulitis, folliculitis, carbuncles, scalded skin syndrome, and abscesses, to life-threatening diseases such as pneumonia, meningitis, osteomyelitis, endocarditis, toxic shock syndrome, bacteremia, and sepsis. It is still one of the five most common causes of hospital-acquired infections and is often the cause of wound infections following surgery. Each year, around 500,000 hospital patients in the United States contract a staphylococcal infection, chiefly by S. aureus.[8] Up to 50,000 deaths each year in the U.S. are linked to staphylococcal infection.[9]
History
Discovery
In 1880, Alexander Ogston, a Scottish surgeon, discovered that Staphylococcus can cause wound infections after noticing groups of bacteria in pus from a surgical abscess during a procedure he was performing. He named it Staphylococcus after its clustered appearance evident under a microscope. Then, in 1884, German scientist Friedrich Julius Rosenbach identified Staphylococcus aureus, discriminating and separating it from Staphylococcus albus, a related bacterium. In the early 1930s, doctors began to use a more streamlined test to detect the presence of an S. aureus infection by the means of coagulase testing, which enables detection of an enzyme produced by the bacterium. Prior to the 1940s, S. aureus infections were fatal in the majority of patients. However, doctors discovered that the use of penicillin could cure S. aureus infections. Unfortunately, by the end of the 1940s, penicillin resistance became widespread amongst this bacterium population and outbreaks of the resistant strain began to occur.[10]
Evolution
Staphylococcus aureus can be sorted into ten dominant human lineages. There are numerous minor lineages as well, but these are not seen in the population as often. Genomes of bacteria within the same lineage are mostly conserved, with the exception of mobile genetic elements. Mobile genetic elements that are common in S. aureus include bacteriophages, pathogenicity islands, plasmids, transposons, and staphylococcal cassette chromosomes. These elements have enabled S. aureus to continually evolve and gain new traits. There is a great deal of genetic variation within the S. aureus species. A study by Fitzgerald et al. (2001) revealed that approximately 22% of the S. aureus genome is non-coding and thus can differ from bacterium to bacterium. An example of this difference is seen in the species' virulence. Only a few strains of S. aureus are associated with infections in humans. This demonstrates that there is a large range of infectious ability within the species.[11]
It has been proposed that one possible reason for the great deal of heterogeneity within the species could be due to its reliance on heterogeneous infections. This occurs when multiple different types of S. aureus cause an infection within a host. The different strains can secrete different enzymes or bring different antibiotic resistances to the group, increasing its pathogenic ability.[12] Thus, there is a need for a large number of mutations and acquisitions of mobile genetic elements.
Another notable evolutionary process within the S. aureus species is its co-evolution with its human hosts. Over time, this parasitic relationship has led to the bacterium's ability to be carried in the nasopharynx of humans without causing symptoms or infection. This allows it to be passed throughout the human population, increasing its fitness as a species.[13] However, only approximately 50% of the human population are carriers of S. aureus, with 20% as continuous carriers and 30% as intermittent. This leads scientists to believe that there are many factors that determine whether S. aureus is carried asymptomatically in humans, including factors that are specific to an individual person. According to a 1995 study by Hofman et al., these factors may include age, sex, diabetes, and smoking. They also determined some genetic variations in humans that lead to an increased ability for S. aureus to colonize, notably a polymorphism in the glucocorticoid receptor gene that results in larger corticosteroid production. In conclusion, there is evidence that any strain of this bacterium can become invasive, as this is highly dependent upon human factors.[14]
Though S. aureus has quick reproductive and micro-evolutionary rates, there are multiple barriers that prevent evolution with the species. One such barrier is AGR, which is a global accessory gene regulator within the bacteria. This such regulator has been linked to the virulence level of the bacteria. Loss of function mutations within this gene have been found to increase the fitness of the bacterium containing it. Thus, S. aureus must make a trade-off to increase their success as a species, exchanging reduced virulence for increased drug resistance. Another barrier to evolution is the Sau1 Type I restriction modification (RM) system. This system exists to protect the bacterium from foreign DNA by digesting it. Exchange of DNA between the same lineage is not blocked, since they have the same enzymes and the RM system does not recognize the new DNA as foreign, but transfer between different lineages is blocked.[12]
Microbiology
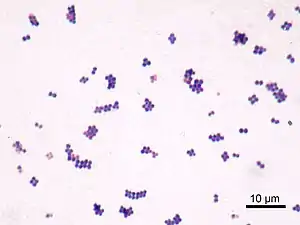

S. aureus (/ˌstæfɪləˈkɒkəs ˈɔːriəs, -loʊ-/,[15][16] Greek σταφυλόκοκκος, "grape-cluster berry", Latin aureus, "golden") is a facultative anaerobic, Gram-positive coccal (round) bacterium also known as "golden staph" and "oro staphira". S. aureus is nonmotile and does not form spores.[17] In medical literature, the bacterium is often referred to as S. aureus, Staph aureus or Staph a..[18] S. aureus appears as staphylococci (grape-like clusters) when viewed through a microscope, and has large, round, golden-yellow colonies, often with hemolysis, when grown on blood agar plates.[19] S. aureus reproduces asexually by binary fission. Complete separation of the daughter cells is mediated by S. aureus autolysin, and in its absence or targeted inhibition, the daughter cells remain attached to one another and appear as clusters.[20]
S. aureus is catalase-positive (meaning it can produce the enzyme catalase). Catalase converts hydrogen peroxide (H
2O
2) to water and oxygen. Catalase-activity tests are sometimes used to distinguish staphylococci from enterococci and streptococci. Previously, S. aureus was differentiated from other staphylococci by the coagulase test. However, not all S. aureus strains are coagulase-positive[19][21] and incorrect species identification can impact effective treatment and control measures.[22]
Natural genetic transformation is a reproductive process involving DNA transfer from one bacterium to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. S. aureus was found to be capable of natural genetic transformation, but only at low frequency under the experimental conditions employed.[23] Further studies suggested that the development of competence for natural genetic transformation may be substantially higher under appropriate conditions, yet to be discovered.[24]
Role in health
In humans, S. aureus can be present in the upper respiratory tract, gut mucosa, and skin as a member of the normal microbiota.[25][26][27] However, because S. aureus can cause disease under certain host and environmental conditions, it is characterized as a "pathobiont".[25]
Role in disease
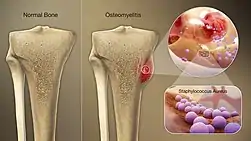

While S. aureus usually acts as a commensal bacterium, asymptomatically colonizing about 30% of the human population, it can sometimes cause disease.[3] In particular, S. aureus is one of the most common causes of bacteremia and infective endocarditis. Additionally, it can cause various skin and soft-tissue infections,[3] particularly when skin or mucosal barriers have been breached.
S. aureus infections can spread through contact with pus from an infected wound, skin-to-skin contact with an infected person, and contact with objects used by an infected person such as towels, sheets, clothing, or athletic equipment. Joint replacements put a person at particular risk of septic arthritis, staphylococcal endocarditis (infection of the heart valves), and pneumonia.[28]
Preventive measures include washing hands, bathing or showering daily, with soap.
S. aureus is a significant cause of chronic biofilm infections on medical implants, and the repressor of toxins is part of the infection pathway.[29]
S. aureus can lay dormant in the body for years undetected. Once symptoms begin to show, the host is contagious for another two weeks, and the overall illness lasts a few weeks. If untreated, though, the disease can be deadly.[30] Deeply penetrating S. aureus infections can be severe.
Skin infections
Skin infections are the most common form of S. aureus infection. This can manifest in various ways, including small benign boils, folliculitis, impetigo, cellulitis, and more severe, invasive soft-tissue infections.[7][3]
S. aureus is extremely prevalent in persons with atopic dermatitis, more commonly known as eczema.[31] It is mostly found in fertile, active places, including the armpits, hair, and scalp. Large pimples that appear in those areas may exacerbate the infection if lacerated. This can lead to staphylococcal scalded skin syndrome, a severe form of which can be seen in newborns.[32]
The presence of S. aureus in persons with atopic dermatitis is not an indication to treat with oral antibiotics, as evidence has not shown this to give benefit to the patient.[33][34] However, topical antibiotics combined with corticosteroids have been found to improve the condition.[35] Colonization of S. aureus drives inflammation of atopic dermatitis;[36][31]S. aureus is believed to exploit defects in the skin barrier of persons with atopic dermatitis, triggering cytokine expression and therefore exacerbating symptoms.[37]
Food poisoning
S. aureus is also responsible for food poisoning and achieves this by generating toxins in the human body.[38] Its incubation period lasts one to six hours,[39] with the illness itself lasting from 30 minutes to 3 days.[40] Preventive measures one can take to help prevent the spread of the disease include washing hands thoroughly with soap and water before preparing food. Stay away from any food if ill, and wear gloves if any open wounds occur on hands or wrists while preparing food. If storing food for longer than 2 hours, keep the food below 4.4 or above 60 °C (below 40 or above 140 °F).[41]
Bone and joint infections
S. aureus is a common cause of major bone and joint infections, including osteomyelitis, septic arthritis, and infections following replacement joint surgeries.[42][3][43]
Bacteremia
S. aureus is a leading cause of bloodstream infections throughout much of the industrialized world.[42] Infection is generally associated with breaks in the skin or mucosal membranes due to surgery, injury, or use of intravascular devices such as catheters, hemodialysis machines, or injected drugs.[3][42] Once the bacteria have entered the bloodstream, they can infect various organs, causing infective endocarditis, septic arthritis, and osteomyelitis.[42] This disease is particularly prevalent and severe in the very young and very old.[3]
Without antibiotic treatment, S. aureus bacteremia has a case fatality rate around 80%.[3] With antibiotic treatment, case fatality rates range from 15% to 50% depending on the age and health of the patient, as well as the antibiotic resistance of the S. aureus strain.[3]
Medical implant infections
S. aureus is often found in biofilms formed on medical devices implanted in the body or on human tissue. It is commonly found with another pathogen, Candida albicans, forming multispecies biofilms. The latter is suspected to help S. aureus penetrate human tissue.[9] A higher mortality is linked with multispecies biofilms.[44]
S. aureus biofilm is the predominant cause of orthopedic implant-related infections, but is also found on cardiac implants, vascular grafts, various catheters, and cosmetic surgical implants.[45][46] After implantation, the surface of these devices becomes coated with host proteins, which provide a rich surface for bacterial attachment and biofilm formation. Once the device becomes infected, it must by completely removed, since S. aureus biofilm cannot be destroyed by antibiotic treatments.[46]
Current therapy for S. aureus biofilm-mediated infections involves surgical removal of the infected device followed by antibiotic treatment. Conventional antibiotic treatment alone is not effective in eradicating such infections.[45] An alternative to postsurgical antibiotic treatment is using antibiotic-loaded, dissolvable calcium sulfate beads, which are implanted with the medical device. These beads can release high doses of antibiotics at the desired site to prevent the initial infection.[46]
Novel treatments for S. aureus biofilm involving nano silver particles, bacteriophages, and plant-derived antibiotic agents are being studied. These agents have shown inhibitory effects against S. aureus embedded in biofilms.[47] A class of enzymes have been found to have biofilm matrix-degrading ability, thus may be used as biofilm dispersal agents in combination with antibiotics.[48]
Animal infections
S. aureus can survive on dogs,[49] cats,[50] and horses,[51] and can cause bumblefoot in chickens.[52] Some believe health-care workers' dogs should be considered a significant source of antibiotic-resistant S. aureus, especially in times of outbreak.[49] In a 2008 study by Boost, O’Donoghue, and James, it was found that just about 90% of S. aureus colonized within pet dogs presented as resistant to at least one antibiotic. The nasal region has been implicated as the most important site of transfer between dogs and humans.[53]
S. aureus is one of the causal agents of mastitis in dairy cows. Its large polysaccharide capsule protects the organism from recognition by the cow's immune defenses.[54]
Virulence factors
Enzymes
S. aureus produces various enzymes such as coagulase (bound and free coagulases) which facilitates the conversion of fibrinogen to fibrin to cause clots which is important in skin infections.[55] Hyaluronidase (also known as spreading factor) breaks down hyaluronic acid and helps in spreading it. Deoxyribonuclease, which breaks down the DNA, protects S. aureus from neutrophil extracellular trap-mediated killing.[56][57] S. aureus also produces lipase to digest lipids, staphylokinase to dissolve fibrin and aid in spread, and beta-lactamase for drug resistance.[58]
Toxins
Depending on the strain, S. aureus is capable of secreting several exotoxins, which can be categorized into three groups. Many of these toxins are associated with specific diseases.[59]
- Superantigens
- Antigens known as superantigens can induce toxic shock syndrome (TSS). This group comprises 25 staphylococcal enterotoxins (SEs) which have been identified to date and named alphabetically (SEA - SEZ),[60] including enterotoxin type B as well as the toxic shock syndrome toxin TSST-1 which causes TSS associated with tampon use. Toxic shock syndrome is characterized by fever, erythematous rash, low blood pressure, shock, multiple organ failure, and skin peeling. Lack of antibody to TSST-1 plays a part in the pathogenesis of TSS. Other strains of S. aureus can produce an enterotoxin that is the causative agent of a type of gastroenteritis. This form of gastroenteritis is self-limiting, characterized by vomiting and diarrhea 1–6 hours after ingestion of the toxin, with recovery in 8 to 24 hours. Symptoms include nausea, vomiting, diarrhea, and major abdominal pain.[61][62]
- Exfoliative toxins
- Exfoliative toxins are exotoxins implicated in the disease staphylococcal scalded skin syndrome (SSSS), which occurs most commonly in infants and young children. It also may occur as epidemics in hospital nurseries. The protease activity of the exfoliative toxins causes peeling of the skin observed with SSSS.[62]
- Other toxins
- Staphylococcal toxins that act on cell membranes include alpha toxin, beta toxin, delta toxin, and several bicomponent toxins. Strains of S. aureus can host phages, such as the prophage Φ-PVL that produces Panton-Valentine leukocidin (PVL), to increase virulence. The bicomponent toxin PVL is associated with severe necrotizing pneumonia in children.[63][64] The genes encoding the components of PVL are encoded on a bacteriophage found in community-associated MRSA strains.
Type VII Secretion system
A secretion system is a highly specialised multi-protein unit that is embedded in the cell envelope with the function of translocating effector proteins from inside of the cell to the extracellular space or into a target host cytosol. The exact structure and function of T7SS is yet to be fully elucidated. Currently, four proteins are known components of S. aureus Type VII Secretion System (T7SS); EssC is a large integral membrane ATPase - which most likely powers the secretion systems and has been hypothesised forming part of the translocation channel. The other proteins are EsaA, EssB, EssA, that are membrane proteins that function alongside EssC to mediate protein secretion. The exact mechanism of how substrates reach the cell surface is unknown, as is the interaction of the three membrane proteins with each other and EssC.[65]
T7 dependent effector proteins
EsaD is DNA endonuclease toxin secreted by S. aureus, has been shown to inhibit growth of competitor S. aureus strain in vitro.[66] EsaD is cosecreted with chaperone EsaE, which stabilises EsaD structure and brings EsaD to EssC for secretion.[66][65] Strains that produce EsaD also co-produce EsaG, a cytoplasmic anti-toxin that protects the producer strain from EsaD's toxicity.[66]
TspA is another toxin that mediates intraspecies competition. It is a bacteriostatic toxin that has a membrane depolarising activity facilitated by its c-terminal domain. Tsai is a transmembrane protein that confers immunity to the producer strain of TspA, as well as the attacked strains. There is genetic variability of the c-terminal domain of TspA therefore, it seems like the strains may produce different TspA variants to increase competitiveness.[67]
Toxins that play a role in intraspecies competition confers an advantage by promoting successful colonisation in polymicrobial communities such as the nasopharynx and lung by outcompeting lesser strains.[67]
There are also T7 effector proteins that play role a in pathogenesis, for example mutational studies of S. aureus have suggested that EsxB and EsxC contribute to persistent infection in a murine abscess model.[68]
EsxX is has been implicated in neutrophil lysis, therefore suggested as contributing to the evasion of host immune system. Deletion of essX in S. aureus resulted in significantly reduced resistance to neutrophils and reduced virulence in murine skin and blood infection models.[69]
Altogether, T7SS and known secreted effector proteins are a strategy of pathogenesis by improving fitness against competitor S. aureus species as well as increased virulence via evading the innate immune system and optimising persistent infections.
Small RNA
The list of small RNAs involved in the control of bacterial virulence in S. aureus is growing. This can be facilitated by factors such as increased biofilm formation in the presence of increased levels of such small RNAs.[70] For example, RNAIII,[71] SprD,[72] SprC,[73][74] RsaE,[75] SprA1,[76] SSR42,[77] ArtR,[78] SprX, and Teg49.[79]
DNA repair
Host neutrophils cause DNA double-strand breaks in S. aureus through the production of reactive oxygen species.[80] For infection of a host to be successful, S. aureus must survive such damages caused by the hosts’ defenses. The two protein complex RexAB encoded by S. aureus is employed in the recombinational repair of DNA double-strand breaks.[80]
Strategies for post-transcriptional regulation by 3'untranslated region
Many mRNAs in S. aureus carry three prime untranslated regions (3'UTR) longer than 100 nucleotides, which may potentially have a regulatory function.[81]
Further investigation of icaR mRNA (mRNA coding for the repressor of the main expolysaccharidic compound of the bacteria biofilm matrix) demonstrated that the 3'UTR binding to the 5' UTR can interfere with the translation initiation complex and generate a double stranded substrate for RNase III. The interaction is between the UCCCCUG motif in the 3'UTR and the Shine-Dalagarno region at the 5'UTR. Deletion of the motif resulted in IcaR repressor accumulation and inhibition of biofilm development.[81] The biofilm formation is the main cause of Staphylococcus implant infections.[82]
Biofilm
Biofilms are groups of microorganisms, such as bacteria, that attach to each other and grow on wet surfaces.[83] The S. aureus biofilm is embedded in a glycocalyx slime layer and can consist of teichoic acids, host proteins, extracellular DNA (eDNA) and sometimes polysaccharide intercellular antigen (PIA). S. aureus biofilms are important in disease pathogenesis, as they can contribute to antibiotic resistance and immune system evasion.[46] S. aureus biofilm has high resistance to antibiotic treatments and host immune response.[83] One hypothesis for explaining this is that the biofilm matrix protects the embedded cells by acting as a barrier to prevent antibiotic penetration. However, the biofilm matrix is composed with many water channels, so this hypothesis is becoming increasingly less likely, but a biofilm matrix possibly contains antibiotic‐degrading enzymes such as β-lactamases, which can prevent antibiotic penetration.[84] Another hypothesis is that the conditions in the biofilm matrix favor the formation of persister cells, which are highly antibiotic-resistant, dormant bacterial cells.[46] S. aureus biofilms also have high resistance to host immune response. Though the exact mechanism of resistance is unknown, S. aureus biofilms have increased growth under the presence of cytokines produced by the host immune response.[85] Host antibodies are less effective for S. aureus biofilm due to the heterogeneous antigen distribution, where an antigen may be present in some areas of the biofilm, but completely absent from other areas.[46]
Studies in biofilm development have shown to be related to changes in gene expression. There are specific genes that were found to be crucial in the different biofilm growth stages. Two of these genes include rocD and gudB, which encode for the enzyme's ornithine-oxo-acid transaminase and glutamate dehydrogenase, which are important for amino acid metabolism. Studies have shown biofilm development rely on amino acids glutamine and glutamate for proper metabolic functions.[86]
Other immunoevasive strategies
- Protein A
Protein A is anchored to staphylococcal peptidoglycan pentaglycine bridges (chains of five glycine residues) by the transpeptidase sortase A.[87] Protein A, an IgG-binding protein, binds to the Fc region of an antibody. In fact, studies involving mutation of genes coding for protein A resulted in a lowered virulence of S. aureus as measured by survival in blood, which has led to speculation that protein A-contributed virulence requires binding of antibody Fc regions.[88]
Protein A in various recombinant forms has been used for decades to bind and purify a wide range of antibodies by immunoaffinity chromatography. Transpeptidases, such as the sortases responsible for anchoring factors like protein A to the staphylococcal peptidoglycan, are being studied in hopes of developing new antibiotics to target MRSA infections.[89]

- Staphylococcal pigments
Some strains of S. aureus are capable of producing staphyloxanthin — a golden-coloured carotenoid pigment. This pigment acts as a virulence factor, primarily by being a bacterial antioxidant which helps the microbe evade the reactive oxygen species which the host immune system uses to kill pathogens.[90][91]
Mutant strains of S. aureus modified to lack staphyloxanthin are less likely to survive incubation with an oxidizing chemical, such as hydrogen peroxide, than pigmented strains. Mutant colonies are quickly killed when exposed to human neutrophils, while many of the pigmented colonies survive.[90] In mice, the pigmented strains cause lingering abscesses when inoculated into wounds, whereas wounds infected with the unpigmented strains quickly heal.
These tests suggest the Staphylococcus strains use staphyloxanthin as a defence against the normal human immune system. Drugs designed to inhibit the production of staphyloxanthin may weaken the bacterium and renew its susceptibility to antibiotics.[91] In fact, because of similarities in the pathways for biosynthesis of staphyloxanthin and human cholesterol, a drug developed in the context of cholesterol-lowering therapy was shown to block S. aureus pigmentation and disease progression in a mouse infection model.[92]
Classical diagnosis

Depending upon the type of infection present, an appropriate specimen is obtained accordingly and sent to the laboratory for definitive identification by using biochemical or enzyme-based tests. A Gram stain is first performed to guide the way, which should show typical Gram-positive bacteria, cocci, in clusters. Second, the isolate is cultured on mannitol salt agar, which is a selective medium with 7.5% NaCl that allows S. aureus to grow, producing yellow-colored colonies as a result of mannitol fermentation and subsequent drop in the medium's pH.[93][94]
Furthermore, for differentiation on the species level, catalase (positive for all Staphylococcus species), coagulase (fibrin clot formation, positive for S. aureus), DNAse (zone of clearance on DNase agar), lipase (a yellow color and rancid odor smell), and phosphatase (a pink color) tests are all done. For staphylococcal food poisoning, phage typing can be performed to determine whether the staphylococci recovered from the food were the source of infection.[95]
Rapid diagnosis and typing
Diagnostic microbiology laboratories and reference laboratories are key for identifying outbreaks and new strains of S. aureus. Recent genetic advances have enabled reliable and rapid techniques for the identification and characterization of clinical isolates of S. aureus in real time. These tools support infection control strategies to limit bacterial spread and ensure the appropriate use of antibiotics. Quantitative PCR is increasingly being used to identify outbreaks of infection.[96][97]
When observing the evolvement of S. aureus and its ability to adapt to each modified antibiotic, two basic methods known as "band-based" or "sequence-based" are employed.[98] Keeping these two methods in mind, other methods such as multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE), bacteriophage typing, spa locus typing, and SCCmec typing are often conducted more than others.[99] With these methods, it can be determined where strains of MRSA originated and also where they are currently.[100]
With MLST, this technique of typing uses fragments of several housekeeping genes known as aroE, glpF, gmk, pta, tip, and yqiL. These sequences are then assigned a number which give to a string of several numbers that serve as the allelic profile. Although this is a common method, a limitation about this method is the maintenance of the microarray which detects newly allelic profiles, making it a costly and time-consuming experiment.[98]
With PFGE, a method which is still very much used dating back to its first success in 1980s, remains capable of helping differentiate MRSA isolates.[100] To accomplish this, the technique uses multiple gel electrophoresis, along with a voltage gradient to display clear resolutions of molecules. The S. aureus fragments then transition down the gel, producing specific band patterns that are later compared with other isolates in hopes of identifying related strains. Limitations of the method include practical difficulties with uniform band patterns and PFGE sensitivity as a whole.
Spa locus typing is also considered a popular technique that uses a single locus zone in a polymorphic region of S. aureus to distinguish any form of mutations.[100] Although this technique is often inexpensive and less time-consuming, the chance of losing discriminatory power making it hard to differentiate between MLST clonal complexes exemplifies a crucial limitation.
Treatment
For susceptible strains, the treatment of choice for S. aureus infection is penicillin. An antibiotic derived from some Penicillium fungal species, penicillin inhibits the formation of peptidoglycan cross-linkages that provide the rigidity and strength in a bacterial cell wall. The four-membered β-lactam ring of penicillin is bound to enzyme DD-transpeptidase, an enzyme that when functional, cross-links chains of peptidoglycan that form bacterial cell walls. The binding of β-lactam to DD-transpeptidase inhibits the enzyme's functionality and it can no longer catalyze the formation of the cross-links. As a result, cell wall formation and degradation are imbalanced, thus resulting in cell death. In most countries, however, penicillin resistance is extremely common (>90%), and first-line therapy is most commonly a penicillinase-resistant β-lactam antibiotic (for example, oxacillin or flucloxacillin, both of which have the same mechanism of action as penicillin) or vancomycin, depending on local resistance patterns. Combination therapy with gentamicin may be used to treat serious infections, such as endocarditis,[101][102] but its use is controversial because of the high risk of damage to the kidneys.[103] The duration of treatment depends on the site of infection and on severity. Adjunctive rifampicin has been historically used in the management of S aureus bacteraemia, but randomised controlled trial evidence has shown this to be of no overall benefit over standard antibiotic therapy.[104]
Antibiotic resistance in S. aureus was uncommon when penicillin was first introduced in 1943. Indeed, the original Petri dish on which Alexander Fleming of Imperial College London observed the antibacterial activity of the Penicillium fungus was growing a culture of S. aureus. By 1950, 40% of hospital S. aureus isolates were penicillin-resistant; by 1960, this had risen to 80%.[105]
Methicillin-resistant Staphylococcus aureus (MRSA, often pronounced /ˈmɜːrsə/ or /ɛm ɑːr ɛs eɪ/), is one of a number of greatly feared strains of S. aureus which have become resistant to most β-lactam antibiotics. For this reason, vancomycin, a glycopeptide antibiotic, is commonly used to combat MRSA. Vancomycin inhibits the synthesis of peptidoglycan, but unlike β-lactam antibiotics, glycopeptide antibiotics target and bind to amino acids in the cell wall, preventing peptidoglycan cross-linkages from forming. MRSA strains are most often found associated with institutions such as hospitals, but are becoming increasingly prevalent in community-acquired infections.
Minor skin infections can be treated with triple antibiotic ointment.[106] One topical agent that is prescribed is Mupirocin, a protein synthesis inhibitor that is produced naturally by Pseudomonas fluorescens and has seen success for treatment of S. aureus nasal carriage.[46]
Antibiotic resistance

S. aureus was found to be the second leading pathogen for deaths associated with antimicrobial resistance in 2019.[107]
Staphylococcal resistance to penicillin is mediated by penicillinase (a form of beta-lactamase) production: an enzyme that cleaves the β-lactam ring of the penicillin molecule, rendering the antibiotic ineffective. Penicillinase-resistant β-lactam antibiotics, such as methicillin, nafcillin, oxacillin, cloxacillin, dicloxacillin, and flucloxacillin are able to resist degradation by staphylococcal penicillinase.
Resistance to methicillin is mediated via the mec operon, part of the staphylococcal cassette chromosome mec (SCCmec). SCCmec is a family of mobile genetic elements, which is a major driving force of S. aureus evolution.[98] Resistance is conferred by the mecA gene, which codes for an altered penicillin-binding protein (PBP2a or PBP2') that has a lower affinity for binding β-lactams (penicillins, cephalosporins, and carbapenems). This allows for resistance to all β-lactam antibiotics, and obviates their clinical use during MRSA infections. Studies have explained that this mobile genetic element has been acquired by different lineages in separate gene transfer events, indicating that there is not a common ancestor of differing MRSA strains.[108] Interestingly, one study suggests that MRSA sacrifices virulence, for example, toxin production and invasiveness, for survival and creation of biofilms[109]
Aminoglycoside antibiotics, such as kanamycin, gentamicin, streptomycin, were once effective against staphylococcal infections until strains evolved mechanisms to inhibit the aminoglycosides' action, which occurs via protonated amine and/or hydroxyl interactions with the ribosomal RNA of the bacterial 30S ribosomal subunit.[110] Three main mechanisms of aminoglycoside resistance mechanisms are currently and widely accepted: aminoglycoside modifying enzymes, ribosomal mutations, and active efflux of the drug out of the bacteria.
Aminoglycoside-modifying enzymes inactivate the aminoglycoside by covalently attaching either a phosphate, nucleotide, or acetyl moiety to either the amine or the alcohol key functional group (or both groups) of the antibiotic. This changes the charge or sterically hinders the antibiotic, decreasing its ribosomal binding affinity. In S. aureus, the best-characterized aminoglycoside-modifying enzyme is aminoglycoside adenylyltransferase 4' IA (ANT(4')IA). This enzyme has been solved by X-ray crystallography.[111] The enzyme is able to attach an adenyl moiety to the 4' hydroxyl group of many aminoglycosides, including kamamycin and gentamicin.
Glycopeptide resistance is mediated by acquisition of the vanA gene, which originates from the Tn1546 transposon found in a plasmid in enterococci and codes for an enzyme that produces an alternative peptidoglycan to which vancomycin will not bind.[112]
Today, S. aureus has become resistant to many commonly used antibiotics. In the UK, only 2% of all S. aureus isolates are sensitive to penicillin, with a similar picture in the rest of the world. The β-lactamase-resistant penicillins (methicillin, oxacillin, cloxacillin, and flucloxacillin) were developed to treat penicillin-resistant S. aureus, and are still used as first-line treatment. Methicillin was the first antibiotic in this class to be used (it was introduced in 1959), but, only two years later, the first case of methicillin-resistant Staphylococcus aureus (MRSA) was reported in England.[113]
Despite this, MRSA generally remained an uncommon finding, even in hospital settings, until the 1990s, when the MRSA prevalence in hospitals exploded, and it is now endemic.[114] Now, methicillin-resistant Staphylococcus aureus (MRSA) is not only a human pathogen causing a variety of infections, such as skin and soft tissue infection (SSTI), pneumonia, and sepsis, but it also can cause disease in animals, known as livestock-associated MRSA (LA-MRSA).[115]
MRSA infections in both the hospital and community setting are commonly treated with non-β-lactam antibiotics, such as clindamycin (a lincosamine) and co-trimoxazole (also commonly known as trimethoprim/sulfamethoxazole). Resistance to these antibiotics has also led to the use of new, broad-spectrum anti-Gram-positive antibiotics, such as linezolid, because of its availability as an oral drug. First-line treatment for serious invasive infections due to MRSA is currently glycopeptide antibiotics (vancomycin and teicoplanin). A number of problems with these antibiotics occur, such as the need for intravenous administration (no oral preparation is available), toxicity, and the need to monitor drug levels regularly by blood tests. Also, glycopeptide antibiotics do not penetrate very well into infected tissues (this is a particular concern with infections of the brain and meninges and in endocarditis). Glycopeptides must not be used to treat methicillin-sensitive S. aureus (MSSA), as outcomes are inferior.[116]
Because of the high level of resistance to penicillins and because of the potential for MRSA to develop resistance to vancomycin, the U.S. Centers for Disease Control and Prevention has published guidelines for the appropriate use of vancomycin. In situations where the incidence of MRSA infections is known to be high, the attending physician may choose to use a glycopeptide antibiotic until the identity of the infecting organism is known. After the infection is confirmed to be due to a methicillin-susceptible strain of S. aureus, treatment can be changed to flucloxacillin or even penicillin, as appropriate.
Vancomycin-resistant S. aureus (VRSA) is a strain of S. aureus that has become resistant to the glycopeptides. The first case of vancomycin-intermediate S. aureus (VISA) was reported in Japan in 1996;[117] but the first case of S. aureus truly resistant to glycopeptide antibiotics was only reported in 2002.[118] Three cases of VRSA infection had been reported in the United States as of 2005.[119] At least in part the antimicrobial resistance in S. aureus can be explained by its ability to adapt. Multiple two component signal transduction pathways helps S. aureus to express genes that are required to survive under antimicrobial stress.[120]
Efflux pumps
Among the various mechanisms that MRSA acquires to elude antibiotic resistance (e.g., drug inactivation, target alteration, reduction of permeability) there is also the overexpression of efflux pumps. Efflux pumps are membrane-integrated proteins that are physiologically needed in the cell for the exportation of xenobiotic compounds. They are divided into six families, each of which has a different structure, function, and transport of energy. The main efflux pumps of S. Aureus are the MFS (Major Facilitator Superfamily) which includes the MdeA pump as well as the NorA pump and the MATE (Multidrug and Toxin Extrusion) to which it belongs the MepA pump. For transport, these families use an electrochemical potential and an ion concentration gradient, while the ATP-binding cassette (ABC) family acquires its energy from the hydrolysis of ATP.
These pumps are overexpressed by MDR S. aureus (Multidrug resistant S. aureus) and the result is an excessive expulsion of the antibiotic outside the cell, which makes its action ineffective. Efflux pumps also contribute significantly to the development of impenetrable biofilms.
By directly modulating efflux pumps' activity or decreasing their expression, it may be possible to modify the resistant phenotype and restore the effectiveness of existing antibiotics.[121]
Carriage
About 33% of the U.S. population are carriers of S. aureus and about 2% carry MRSA.[122] Even healthcare providers can be MRSA colonizers.[123][124]
The carriage of S. aureus is an important source of hospital-acquired infection (also called nosocomial) and community-acquired MRSA. Although S. aureus can be present on the skin of the host, a large proportion of its carriage is through the anterior nares of the nasal passages[2] and can further be present in the ears.[125] The ability of the nasal passages to harbour S. aureus results from a combination of a weakened or defective host immunity and the bacterium's ability to evade host innate immunity.[126] Nasal carriage is also implicated in the occurrence of staph infections.[127]
Infection control
Spread of S. aureus (including MRSA) generally is through human-to-human contact, although recently some veterinarians have discovered the infection can be spread through pets,[128] with environmental contamination thought to play a relatively less important part.[129] Emphasis on basic hand washing techniques are, therefore, effective in preventing its transmission. The use of disposable aprons and gloves by staff reduces skin-to-skin contact, so further reduces the risk of transmission.
Recently, myriad cases of S. aureus have been reported in hospitals across America. Transmission of the pathogen is facilitated in medical settings where healthcare worker hygiene is insufficient. S. aureus is an incredibly hardy bacterium, as was shown in a study where it survived on polyester for just under three months;[130] polyester is the main material used in hospital privacy curtains.
The bacteria are transported on the hands of healthcare workers, who may pick them up from a seemingly healthy patient carrying a benign or commensal strain of S. aureus, and then pass it on to the next patient being treated. Introduction of the bacteria into the bloodstream can lead to various complications, including endocarditis, meningitis, and, if it is widespread, sepsis.
Ethanol has proven to be an effective topical sanitizer against MRSA. Quaternary ammonium can be used in conjunction with ethanol to increase the duration of the sanitizing action. The prevention of nosocomial infections involves routine and terminal cleaning. Nonflammable alcohol vapor in CO
2 NAV-CO2 systems have an advantage, as they do not attack metals or plastics used in medical environments, and do not contribute to antibacterial resistance.
An important and previously unrecognized means of community-associated MRSA colonization and transmission is during sexual contact.[131]
S. aureus is killed in one minute at 78 °C and in ten minutes at 64 °C but is resistant to freezing.[132][133]
Certain strains of S. aureus have been described as being resistant to chlorine disinfection.[134][135]
The use of mupirocin ointment can reduce the rate of infections due to nasal carriage of S. aureus.[136] There is limited evidence that nasal decontamination of S. aureus using antibiotics or antiseptics can reduce the rates of surgical site infections.[137]
| Top common bacterium in each industry | |
|---|---|
| Catering industry | Vibrio parahaemolyticus, S. aureus, Bacillus cereus |
| Medical industry | Escherichia coli, S. aureus, Pseudomonas aeruginosa[138] |
Research
As of 2021, no approved vaccine exists against S. aureus. Early clinical trials have been conducted for several vaccines candidates such as Nabi's StaphVax and PentaStaph, Intercell's / Merck's V710, VRi's SA75, and others.[139]
While some of these vaccines candidates have shown immune responses, others aggravated an infection by S. aureus. To date, none of these candidates provides protection against a S. aureus infection. The development of Nabi's StaphVax was stopped in 2005 after phase III trials failed.[140] Intercell's first V710 vaccine variant was terminated during phase II/III after higher mortality and morbidity were observed among patients who developed S. aureus infection.[141]
Nabi's enhanced S. aureus vaccines candidate PentaStaph was sold in 2011 to GlaxoSmithKline Biologicals S.A.[142] The current status of PentaStaph is unclear. A WHO document indicates that PentaStaph failed in the phase III trial stage.[143]
In 2010, GlaxoSmithKline started a phase 1 blind study to evaluate its GSK2392103A vaccine.[144] As of 2016, this vaccine is no longer under active development.[145]
Pfizer's S. aureus four-antigen vaccine SA4Ag was granted fast track designation by the U.S. Food and Drug Administration in February 2014.[146] In 2015, Pfizer has commenced a phase 2b trial regarding the SA4Ag vaccine.[147] Phase 1 results published in February 2017 showed a very robust and secure immunogenicity of SA4Ag.[148] The vaccine underwent clinical trial until June 2019, with results published in September 2020, that did not demonstrate a significant reduction in Postoperative Bloodstream Infection after Surgery.[147]
In 2015, Novartis Vaccines and Diagnostics, a former division of Novartis and now part of GlaxoSmithKline, published promising pre-clinical results of their four-component Staphylococcus aureus vaccine, 4C-staph.[149]
Standard strains
A number of standard strains of S. aureus (called "type cultures") are used in research and in laboratory testing, such as:
| Name | NCTC | ATCC | Year of deposit | Comment |
|---|---|---|---|---|
| Oxford H | 6571 | 9144 | 1943 | Standard strain used for testing penicillin potency and by which the penicillin unit was originally defined.[150][151] |
| Rosenbach | 12973 | 29213 | 1884 | Standard strain for EUCAST antimicrobial resistance testing.[152] |
See also
- Bundaberg tragedy, deaths of 12 children inoculated with an S. aureus-contaminated vaccine
References
- Masalha M, Borovok I, Schreiber R, Aharonowitz Y, Cohen G (December 2001). "Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen". Journal of Bacteriology. 183 (24): 7260–7272. doi:10.1128/JB.183.24.7260-7272.2001. PMC 95576. PMID 11717286.
- Kluytmans J, van Belkum A, Verbrugh H (July 1997). "Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks". Clinical Microbiology Reviews. 10 (3): 505–520. doi:10.1128/CMR.10.3.505. PMC 172932. PMID 9227864.
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG (July 2015). "Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management". Clinical Microbiology Reviews. 28 (3): 603–661. doi:10.1128/CMR.00134-14. PMC 4451395. PMID 26016486.
- Cole AM, Tahk S, Oren A, Yoshioka D, Kim YH, Park A, Ganz T (November 2001). "Determinants of Staphylococcus aureus nasal carriage". Clinical and Diagnostic Laboratory Immunology. 8 (6): 1064–1069. doi:10.1128/CDLI.8.6.1064-1069.2001. PMC 96227. PMID 11687441.
- Senok AC, Verstraelen H, Temmerman M, Botta GA (October 2009). "Probiotics for the treatment of bacterial vaginosis". The Cochrane Database of Systematic Reviews (4): CD006289. doi:10.1002/14651858.CD006289.pub2. PMID 19821358.
- Hoffman B (2012). Williams gynecology (2nd ed.). New York: McGraw-Hill Medical. p. 65. ISBN 978-0071716727.
- "Staphylococcal Infections". MedlinePlus [Internet]. Bethesda, MD: National Library of Medicine, US.
Skin infections are the most common. They can look like pimples or boils.
- Bowersox J (27 May 1999). "Experimental Staph Vaccine Broadly Protective in Animal Studies". NIH. Archived from the original on 5 May 2007. Retrieved 28 July 2007.
- Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, Filler SG, et al. (January 2015). "Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue". Microbiology. 161 (Pt 1): 168–181. doi:10.1099/mic.0.083485-0. PMC 4274785. PMID 25332378.
- Orent W (2006). "A Brief History of Staph". Proto Magazine.
- Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM (July 2001). "Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic". Proceedings of the National Academy of Sciences of the United States of America. 98 (15): 8821–8826. Bibcode:2001PNAS...98.8821F. doi:10.1073/pnas.161098098. PMC 37519. PMID 11447287.
- Lindsay JA (February 2010). "Genomic variation and evolution of Staphylococcus aureus". International Journal of Medical Microbiology. 300 (2–3): 98–103. doi:10.1016/j.ijmm.2009.08.013. PMID 19811948.
- Fitzgerald JR (January 2014). "Evolution of Staphylococcus aureus during human colonization and infection". Infection, Genetics and Evolution. 21: 542–547. doi:10.1016/j.meegid.2013.04.020. PMID 23624187.
- van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, et al. (January 2009). "Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus". Infection, Genetics and Evolution. 9 (1): 32–47. doi:10.1016/j.meegid.2008.09.012. PMID 19000784.
- "Staphylococcus". Dictionary.com Unabridged (Online). n.d. "aureus". Dictionary.com Unabridged (Online). n.d.
- "staphylococcus – definition of staphylococcus in English from the Oxford dictionary". OxfordDictionaries.com. Archived from the original on 18 August 2012. Retrieved 20 January 2016. "aureus – definition of aureus in English from the Oxford dictionary". OxfordDictionaries.com. Archived from the original on 16 July 2012. Retrieved 20 January 2016.
- "PATHOGEN SAFETY DATA SHEET – INFECTIOUS SUBSTANCES." Staphylococcus cells have a diameter of 0.7–1.2 um. Staphylococcus Aureus. Public Health Agency of Canada, 2011. Web
- "Canadian Centre for Occupational Health and Safety". Retrieved 8 April 2016.
- Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 978-0-8385-8529-0.
- Varrone JJ, de Mesy Bentley KL, Bello-Irizarry SN, Nishitani K, Mack S, Hunter JG, et al. (October 2014). "Passive immunization with anti-glucosaminidase monoclonal antibodies protects mice from implant-associated osteomyelitis by mediating opsonophagocytosis of Staphylococcus aureus megaclusters". Journal of Orthopaedic Research. 32 (10): 1389–1396. doi:10.1002/jor.22672. PMC 4234088. PMID 24992290.
- PreTest, Surgery, 12th ed., p.88
- Matthews KR, Roberson J, Gillespie BE, Luther DA, Oliver SP (June 1997). "Identification and Differentiation of Coagulase-Negative Staphylococcus aureus by Polymerase Chain Reaction". Journal of Food Protection. 60 (6): 686–688. doi:10.4315/0362-028X-60.6.686. PMID 31195568.
- Morikawa K, Takemura AJ, Inose Y, Tsai M, Nguyen TL, Ohta T, Msadek T (2012). "Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus". PLOS Pathogens. 8 (11): e1003003. doi:10.1371/journal.ppat.1003003. PMC 3486894. PMID 23133387.
- Fagerlund A, Granum PE, Håvarstein LS (November 2014). "Staphylococcus aureus competence genes: mapping of the SigH, ComK1 and ComK2 regulons by transcriptome sequencing". Molecular Microbiology. 94 (3): 557–579. doi:10.1111/mmi.12767. PMID 25155269. S2CID 1568023.
- Schenck LP, Surette MG, Bowdish DM (November 2016). "Composition and immunological significance of the upper respiratory tract microbiota". FEBS Letters. 590 (21): 3705–3720. doi:10.1002/1873-3468.12455. PMC 7164007. PMID 27730630.
- Wollina U (2017). "Microbiome in atopic dermatitis". Clinical, Cosmetic and Investigational Dermatology. 10: 51–56. doi:10.2147/CCID.S130013. PMC 5327846. PMID 28260936.
- Otto M (April 2010). "Staphylococcus colonization of the skin and antimicrobial peptides". Expert Review of Dermatology. 5 (2): 183–195. doi:10.1586/edm.10.6. PMC 2867359. PMID 20473345.
- Kuehnert MJ, Hill HA, Kupronis BA, Tokars JI, Solomon SL, Jernigan DB (June 2005). "Methicillin-resistant-Staphylococcus aureus hospitalizations, United States". Emerging Infectious Diseases. 11 (6): 868–872. doi:10.3201/eid1106.040831. PMC 3367609. PMID 15963281.
- Kavanaugh JS, Horswill AR (June 2016). "Impact of Environmental Cues on Staphylococcal Quorum Sensing and Biofilm Development". The Journal of Biological Chemistry (Review). 291 (24): 12556–12564. doi:10.1074/jbc.R116.722710. PMC 4933443. PMID 27129223.
- "Staphylococcus aureus in Healthcare Settings | HAI". CDC. Retrieved 19 April 2017.
- Monnot GC, Wegrecki M, Cheng TY, Chen YL, Sallee BN, Chakravarthy R, Karantza IM, Tin SY, Khaleel AE, Monga I, Uwakwe LN, Tillman A, Cheng B, Youssef S, Ng SW, Shahine A, Garcia-Vilas JA, Uhlemann AC, Bordone LA, Han A, Rohde CH, Ogg G, Moody DB, Rossjohn J, de Jong A (January 2023). "Staphylococcal phosphatidylglycerol antigens activate human T cells via CD1a". Nature Immunology. 24 (1): 110–122. doi:10.1038/s41590-022-01375-z. PMC 10389259. PMID 35265979. S2CID 255039948.
- Curran JP, Al-Salihi FL (August 1980). "Neonatal staphylococcal scalded skin syndrome: massive outbreak due to an unusual phage type". Pediatrics. 66 (2): 285–290. doi:10.1542/peds.66.2.285. PMID 6447271. S2CID 21783186.
- American Academy of Dermatology (February 2013), "Five Things Physicians and Patients Should Question", Choosing Wisely: an initiative of the ABIM Foundation, American Academy of Dermatology, retrieved 5 December 2013
- George SM, Karanovic S, Harrison DA, Rani A, Birnie AJ, Bath-Hextall FJ, et al. (October 2019). "Interventions to reduce Staphylococcus aureus in the management of eczema". The Cochrane Database of Systematic Reviews. 2019 (10). doi:10.1002/14651858.CD003871.pub3. PMC 6818407. PMID 31684694.
- Hon KL, Wang SS, Lee KK, Lee VW, Leung TF, Ip M (July 2012). "Combined antibiotic/corticosteroid cream in the empirical treatment of moderate to severe eczema: friend or foe?". Journal of Drugs in Dermatology. 11 (7): 861–864. PMID 22777230.
- Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, et al. (April 2015). "Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis". Immunity. 42 (4): 756–766. doi:10.1016/j.immuni.2015.03.014. PMC 4407815. PMID 25902485.
- Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. (November 2016). "Staphylococcus aureus Exploits Epidermal Barrier Defects in Atopic Dermatitis to Trigger Cytokine Expression". The Journal of Investigative Dermatology. 136 (11): 2192–2200. doi:10.1016/j.jid.2016.05.127. PMC 5103312. PMID 27381887.
- "Staphylococcal Food Poisoning". cdc.gov. hhs.gov. 4 October 2016. Retrieved 23 October 2016.
- "Staphylococcus." Foodsafety.gov, U.S. Department of Health and Human Services, https://www.foodsafety.gov/poisoning/causes/bacteriaviruses/staphylococcus/.
- "Staphylococcal Food Poisoning." Food Safety, Centers for Disease Control and Prevention, 4 October 2016, https://www.cdc.gov/foodsafety/diseases/staphylococcal.html.
- Woodson J. "Centers for disease control and prevention". Food Safety. Archived from the original on 8 February 2016. Retrieved 24 October 2017.
- Rasmussen RV, Fowler VG, Skov R, Bruun NE (January 2011). "Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA". Future Microbiology. 6 (1): 43–56. doi:10.2217/fmb.10.155. PMC 3031962. PMID 21162635.
- Latha T, Anil B, Manjunatha H, Chiranjay M, Elsa D, Baby N, Anice G. MRSA: the leading pathogen of orthopedic infection in a tertiary care hospital, South India. African health sciences. 2019 Apr 16;19(1):1393-401.
- Zago CE, Silva S, Sanitá PV, Barbugli PA, Dias CM, Lordello VB, Vergani CE (2015). "Dynamics of biofilm formation and the interaction between Candida albicans and methicillin-susceptible (MSSA) and -resistant Staphylococcus aureus (MRSA)". PLOS ONE. 10 (4): e0123206. Bibcode:2015PLoSO..1023206Z. doi:10.1371/journal.pone.0123206. PMC 4395328. PMID 25875834.
- Nandakumar V, Chittaranjan S, Kurian VM, Doble M (2013). "Characteristics of bacterial biofilm associated with implant material in clinical practice". Polymer Journal. 45 (2): 137–152. doi:10.1038/pj.2012.130.
- Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME (1 September 2011). "Staphylococcus aureus biofilms: properties, regulation, and roles in human disease". Virulence. 2 (5): 445–459. doi:10.4161/viru.2.5.17724. PMC 3322633. PMID 21921685.
- Chung PY, Toh YS (April 2014). "Anti-biofilm agents: recent breakthrough against multi-drug resistant Staphylococcus aureus". Pathogens and Disease. 70 (3): 231–239. doi:10.1111/2049-632x.12141. PMID 24453168.
- Hogan S, Zapotoczna M, Stevens NT, Humphreys H, O'Gara JP, O'Neill E (June 2017). "Potential use of targeted enzymatic agents in the treatment of Staphylococcus aureus biofilm-related infections". The Journal of Hospital Infection. 96 (2): 177–182. doi:10.1016/j.jhin.2017.02.008. PMID 28351512.
- Boost MV, O'Donoghue MM, James A (July 2008). "Prevalence of Staphylococcus aureus carriage among dogs and their owners". Epidemiology and Infection. 136 (7): 953–964. doi:10.1017/S0950268807009326. PMC 2870875. PMID 17678561.
- Hanselman BA, Kruth SA, Rousseau J, Weese JS (September 2009). "Coagulase positive staphylococcal colonization of humans and their household pets". The Canadian Veterinary Journal. 50 (9): 954–958. PMC 2726022. PMID 19949556.
- Burton S, Reid-Smith R, McClure JT, Weese JS (August 2008). "Staphylococcus aureus colonization in healthy horses in Atlantic Canada". The Canadian Veterinary Journal. 49 (8): 797–799. PMC 2465786. PMID 18978975.
- "Staphylococcosis, Staphylococcal Arthritis, Bumble Foot". The Poultry Site. Retrieved 22 October 2013.
- Boost MV, O'Donoghue MM, James A (July 2008). "Prevalence of Staphylococcus aureus carriage among dogs and their owners". Epidemiology and Infection. 136 (7): 953–964. doi:10.1017/s0950268807009326. hdl:10397/7558. PMC 2870875. PMID 17678561.
- Cenci-Goga BT, Karama M, Rossitto PV, Morgante RA, Cullor JS (September 2003). "Enterotoxin production by Staphylococcus aureus isolated from mastitic cows". Journal of Food Protection. 66 (9): 1693–1696. doi:10.4315/0362-028X-66.9.1693. PMID 14503727.
- Cheung GY, Bae JS, Otto M (December 2021). "Pathogenicity and virulence of Staphylococcus aureus". Virulence. 12 (1): 547–569. doi:10.1080/21505594.2021.1878688. PMC 7872022. PMID 33522395.
- Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M (2010). "Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps". Journal of Innate Immunity. 2 (6): 576–586. doi:10.1159/000319909. PMC 2982853. PMID 20829609.
- Monteith AJ, Miller JM, Maxwell CN, Chazin WJ, Skaar EP (September 2021). "Neutrophil extracellular traps enhance macrophage killing of bacterial pathogens". Science Advances. 7 (37): eabj2101. Bibcode:2021SciA....7.2101M. doi:10.1126/sciadv.abj2101. PMC 8442908. PMID 34516771.
- Medical Laboratory Manual For Tropical Countries vol two
- Dinges MM, Orwin PM, Schlievert PM (January 2000). "Exotoxins of Staphylococcus aureus". Clinical Microbiology Reviews. 13 (1): 16–34, table of contents. doi:10.1128/cmr.13.1.16. PMC 88931. PMID 10627489.
- Etter D, Schelin J, Schuppler M, Johler S (September 2020). "Staphylococcal Enterotoxin C-An Update on SEC Variants, Their Structure and Properties, and Their Role in Foodborne Intoxications". Toxins. 12 (9): 584. doi:10.3390/toxins12090584. PMC 7551944. PMID 32927913.
- Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, et al. (January 2001). "egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus". Journal of Immunology. 166 (1): 669–677. doi:10.4049/jimmunol.166.1.669. PMID 11123352.
- Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, Von Eiff C (April 2003). "Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens". Journal of Clinical Microbiology. 41 (4): 1434–1439. doi:10.1128/jcm.41.4.1434-1439.2003. PMC 153929. PMID 12682126.
- Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. (November 1999). "Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia". Clinical Infectious Diseases. 29 (5): 1128–1132. doi:10.1086/313461. PMID 10524952.
- Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M, et al. (March 2002). "Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients". Lancet. 359 (9308): 753–759. doi:10.1016/S0140-6736(02)07877-7. PMID 11888586. S2CID 20400336. As PDF Archived 14 July 2014 at the Wayback Machine
- Bowman L, Palmer T (October 2021). "The Type VII Secretion System of Staphylococcus". Annual Review of Microbiology. 75 (1): 471–494. doi:10.1146/annurev-micro-012721-123600. PMID 34343022. S2CID 236915377.
- Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T (October 2016). "The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria". Nature Microbiology. 2 (1): 16183. doi:10.1038/nmicrobiol.2016.183. PMC 5325307. PMID 27723728.
- Ulhuq FR, Gomes MC, Duggan GM, Guo M, Mendonca C, Buchanan G, et al. (August 2020). "A membrane-depolarizing toxin substrate of the Staphylococcus aureus type VII secretion system mediates intraspecies competition". Proceedings of the National Academy of Sciences of the United States of America. 117 (34): 20836–20847. Bibcode:2020PNAS..11720836U. doi:10.1073/pnas.2006110117. PMC 7456083. PMID 32769205.
- Burts ML, Williams WA, DeBord K, Missiakas DM (January 2005). "EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections". Proceedings of the National Academy of Sciences of the United States of America. 102 (4): 1169–1174. Bibcode:2005PNAS..102.1169B. doi:10.1073/pnas.0405620102. PMC 545836. PMID 15657139.
- Dai Y, Wang Y, Liu Q, Gao Q, Lu H, Meng H, et al. (5 May 2017). "A Novel ESAT-6 Secretion System-Secreted Protein EsxX of Community-Associated Staphylococcus aureus Lineage ST398 Contributes to Immune Evasion and Virulence". Frontiers in Microbiology. 8: 819. doi:10.3389/fmicb.2017.00819. PMC 5418362. PMID 28529509.
- Kim S, Reyes D, Beaume M, Francois P, Cheung A (October 2014). "Contribution of teg49 small RNA in the 5' upstream transcriptional region of sarA to virulence in Staphylococcus aureus". Infection and Immunity. 82 (10): 4369–4379. doi:10.1128/iai.02002-14. PMC 4187880. PMID 25092913.
- Chevalier C, Boisset S, Romilly C, Masquida B, Fechter P, Geissmann T, et al. (March 2010). "Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation". PLOS Pathogens. 6 (3): e1000809. doi:10.1371/journal.ppat.1000809. PMC 2837412. PMID 20300607.
- Chabelskaya S, Gaillot O, Felden B (June 2010). "A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule". PLOS Pathogens. 6 (6): e1000927. doi:10.1371/journal.ppat.1000927. PMC 2880579. PMID 20532214.
- Le Pabic H, Germain-Amiot N, Bordeau V, Felden B (October 2015). "A bacterial regulatory RNA attenuates virulence, spread and human host cell phagocytosis". Nucleic Acids Research. 43 (19): 9232–9248. doi:10.1093/nar/gkv783. PMC 4627067. PMID 26240382.
- Mauro T, Rouillon A, Felden B (December 2016). "Insights into the regulation of small RNA expression: SarA represses the expression of two sRNAs in Staphylococcus aureus". Nucleic Acids Research. 44 (21): 10186–10200. doi:10.1093/nar/gkw777. PMC 5137438. PMID 27596601.
- Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, et al. (October 2010). "Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism". Nucleic Acids Research. 38 (19): 6620–6636. doi:10.1093/nar/gkq462. PMC 2965222. PMID 20511587.
- Sayed N, Jousselin A, Felden B (December 2011). "A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide" (PDF). Nature Structural & Molecular Biology. 19 (1): 105–112. doi:10.1038/nsmb.2193. PMID 22198463. S2CID 8217681.
- Morrison JM, Miller EW, Benson MA, Alonzo F, Yoong P, Torres VJ, et al. (June 2012). "Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative". Journal of Bacteriology. 194 (11): 2924–2938. doi:10.1128/JB.06708-11. PMC 3370614. PMID 22493015.
- Xue T, Zhang X, Sun H, Sun B (February 2014). "ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5' UTR of sarT mRNA". Medical Microbiology and Immunology. 203 (1): 1–12. doi:10.1007/s00430-013-0307-0. PMID 23955428. S2CID 18371872.
- Kim S, Reyes D, Beaume M, Francois P, Cheung A (October 2014). "Contribution of teg49 small RNA in the 5' upstream transcriptional region of sarA to virulence in Staphylococcus aureus". Infection and Immunity. 82 (10): 4369–4379. doi:10.1128/IAI.02002-14. PMC 4187880. PMID 25092913.
- Ha KP, Clarke RS, Kim GL, Brittan JL, Rowley JE, Mavridou DAI, Parker D, Clarke TB, Nobbs AH, Edwards AM. Staphylococcal DNA Repair Is Required for Infection. mBio. 2020 Nov 17;11(6):e02288-20. doi: 10.1128/mBio.02288-20. PMID: 33203752; PMCID: PMC7683395
- Ruiz de los Mozos I, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, et al. (2013). "Base pairing interaction between 5'- and 3'-UTRs controls icaR mRNA translation in Staphylococcus aureus". PLOS Genetics. 9 (12): e1004001. doi:10.1371/journal.pgen.1004001. PMC 3868564. PMID 24367275.
- Arciola CR, Campoccia D, Speziale P, Montanaro L, Costerton JW (September 2012). "Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials". Biomaterials. 33 (26): 5967–5982. doi:10.1016/j.biomaterials.2012.05.031. PMID 22695065.
- Vidyasagar, A. (2016). What Are Biofilms? Live Science.
- de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock RE (October 2013). "Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies". Current Opinion in Microbiology. 16 (5): 580–589. doi:10.1016/j.mib.2013.06.013. PMID 23880136.
- McLaughlin RA, Hoogewerf AJ (August 2006). "Interleukin-1beta-induced growth enhancement of Staphylococcus aureus occurs in biofilm but not planktonic cultures". Microbial Pathogenesis. 41 (2–3): 67–79. doi:10.1016/j.micpath.2006.04.005. PMID 16769197.
- Nassar R, Hachim M, Nassar M, Kaklamanos EG, Jamal M, Williams D, Senok A (2021). "Microbial Metabolic Genes Crucial for S. aureus Biofilms: An Insight From Re-analysis of Publicly Available Microarray Datasets". Frontiers in Microbiology. 11: 607002. doi:10.3389/fmicb.2020.607002. PMC 7876462. PMID 33584569.
- Schneewind O, Fowler A, Faull KF (April 1995). "Structure of the cell wall anchor of surface proteins in Staphylococcus aureus". Science. 268 (5207): 103–106. Bibcode:1995Sci...268..103S. doi:10.1126/science.7701329. PMID 7701329.
- Patel AH, Nowlan P, Weavers ED, Foster T (December 1987). "Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement". Infection and Immunity. 55 (12): 3103–3110. doi:10.1128/IAI.55.12.3103-3110.1987. PMC 260034. PMID 3679545.
- Zhu J, Lu C, Standland M, Lai E, Moreno GN, Umeda A, et al. (February 2008). "Single mutation on the surface of Staphylococcus aureus Sortase A can disrupt its dimerization". Biochemistry. 47 (6): 1667–1674. doi:10.1021/bi7014597. PMID 18193895.
- Clauditz A, Resch A, Wieland KP, Peschel A, Götz F (August 2006). "Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress". Infection and Immunity. 74 (8): 4950–4953. doi:10.1128/IAI.00204-06. PMC 1539600. PMID 16861688.
- Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, et al. (July 2005). "Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity". The Journal of Experimental Medicine. 202 (2): 209–215. doi:10.1084/jem.20050846. PMC 2213009. PMID 16009720.
- Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, et al. (March 2008). "A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence". Science. 319 (5868): 1391–1394. Bibcode:2008Sci...319.1391L. doi:10.1126/science.1153018. PMC 2747771. PMID 18276850.
- Shields, Patricia; Tsang, Anne Y. (9 October 2006). "Mannitol Salt Agar Plates Protocols". www.asmscience.org. Retrieved 31 December 2020.
- "Mannitol Salt Agar (MSA) | Culture Media". Microbe Notes. 14 January 2020. Retrieved 31 December 2020.
- Saint-Martin M, Charest G, Desranleau JM (September 1951). "Bacteriophage typing in investigations of staphylococcal food-poisoning outbreaks". Canadian Journal of Public Health. 42 (9): 351–358. JSTOR 41980177. PMID 14879282.
- Francois P, Schrenzel J (2008). "Rapid Diagnosis and Typing of Staphylococcus aureus". Staphylococcus: Molecular Genetics. Caister Academic Press. ISBN 978-1-904455-29-5.
- Mackay IM, ed. (2007). Real-Time PCR in Microbiology: From Diagnosis to Characterization. Caister Academic Press. ISBN 978-1-904455-18-9.
- Deurenberg RH, Stobberingh EE (December 2008). "The evolution of Staphylococcus aureus". Infection, Genetics and Evolution. 8 (6): 747–763. doi:10.1016/j.meegid.2008.07.007. PMID 18718557.
- Aires de Sousa M, Conceição T, Simas C, de Lencastre H (October 2005). "Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community". Journal of Clinical Microbiology. 43 (10): 5150–5157. doi:10.1128/JCM.43.10.5150-5157.2005. PMC 1248511. PMID 16207977.
- Kim J (2009). "Understanding the Evolution of Methicillin-Resistant Staphylococcus aureus". Clinical Microbiology Newsletter. 31 (3): 17–23. doi:10.1016/j.clinmicnews.2009.01.002.
- Korzeniowski O, Sande MA (October 1982). "Combination antimicrobial therapy for Staphylococcus aureus endocarditis in patients addicted to parenteral drugs and in nonaddicts: A prospective study". Annals of Internal Medicine. 97 (4): 496–503. doi:10.7326/0003-4819-97-4-496. PMID 6751182.
- Bayer AS, Bolger AF, Taubert KA, Wilson W, Steckelberg J, Karchmer AW, et al. (1998). "Diagnosis and management of infective endocarditis and its complications". Circulation. 98 (25): 2936–2948. doi:10.1161/01.CIR.98.25.2936. PMID 9860802.
- Cosgrove SE, Vigliani GA, Fowler VG, Abrutyn E, Corey GR, Levine DP, et al. (March 2009). "Initial low-dose gentamicin for Staphylococcus aureus bacteremia and endocarditis is nephrotoxic". Clinical Infectious Diseases. 48 (6): 713–721. doi:10.1086/597031. PMID 19207079.
- Thwaites GE, Scarborough M, Szubert A, Nsutebu E, Tilley R, Greig J, et al. (February 2018). "Adjunctive rifampicin for Staphylococcus aureus bacteraemia (ARREST): a multicentre, randomised, double-blind, placebo-controlled trial". Lancet. 391 (10121): 668–678. doi:10.1016/S0140-6736(17)32456-X. PMC 5820409. PMID 29249276.
- Chambers HF (2001). "The changing epidemiology of Staphylococcus aureus?". Emerging Infectious Diseases. 7 (2): 178–182. doi:10.3201/eid0702.010204. PMC 2631711. PMID 11294701.
- Bonomo RA, Van Zile PS, Li Q, Shermock KM, McCormick WG, Kohut B (October 2007). "Topical triple-antibiotic ointment as a novel therapeutic choice in wound management and infection prevention: a practical perspective". Expert Review of Anti-Infective Therapy. 5 (5): 773–782. doi:10.1586/14787210.5.5.773. PMID 17914912. S2CID 31594289.
- Murray, Christopher JL; et al. (February 2022). "Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis". Lancet. 399 (10325): 629–655. doi:10.1016/S0140-6736(21)02724-0. PMC 8841637. PMID 35065702.
- Jamrozy D, Coll F, Mather AE, Harris SR, Harrison EM, MacGowan A, et al. (September 2017). "Evolution of mobile genetic element composition in an epidemic methicillin-resistant Staphylococcus aureus: temporal changes correlated with frequent loss and gain events". BMC Genomics. 18 (1): 684. doi:10.1186/s12864-017-4065-z. PMC 5584012. PMID 28870171.
- Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, et al. (5 April 2012). Sullam PM (ed.). "Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections". PLOS Pathogens. 8 (4): e1002626. doi:10.1371/journal.ppat.1002626. PMC 3320603. PMID 22496652.
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V (September 2000). "Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics". Nature. 407 (6802): 340–348. Bibcode:2000Natur.407..340C. doi:10.1038/35030019. PMID 11014183. S2CID 4408938.
- Sakon J, Liao HH, Kanikula AM, Benning MM, Rayment I, Holden HM (November 1993). "Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-A resolution". Biochemistry. 32 (45): 11977–11984. doi:10.1021/bi00096a006. PMID 8218273.
- Arthur M, Courvalin P (August 1993). "Genetics and mechanisms of glycopeptide resistance in enterococci". Antimicrobial Agents and Chemotherapy. ASM. 37 (8): 1563–1571. doi:10.1128/AAC.37.8.1563. PMC 188020. PMID 8215264.
- Rolinson GN, Stevens S, Batchelor FR, Wood JC, Chain EB (September 1960). "Bacteriological studies on a new penicillin-BRL. 1241". Lancet. 2 (7150): 564–567. doi:10.1136/bmj.1.5219.124-a. PMC 1952878. PMID 14438510.
- Johnson AP, Aucken HM, Cavendish S, Ganner M, Wale MC, Warner M, et al. (July 2001). "Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS)". The Journal of Antimicrobial Chemotherapy. 48 (1): 143–144. doi:10.1093/jac/48.1.143. PMID 11418528.
- Chen CJ, Huang YC (August 2018). "Emergence of livestock-associated methicillin-resistant Staphylococcus aureus: Should it be a concern?". Journal of the Formosan Medical Association = Taiwan Yi Zhi. 117 (8): 658–661. doi:10.1016/j.jfma.2018.04.004. PMID 29754805. S2CID 21659477.
- Blot SI, Vandewoude KH, Hoste EA, Colardyn FA (October 2002). "Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus". Archives of Internal Medicine. 162 (19): 2229–2235. doi:10.1001/archinte.162.19.2229. PMID 12390067.
- Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC (July 1997). "Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility". The Journal of Antimicrobial Chemotherapy. 40 (1): 135–136. doi:10.1093/jac/40.1.135. PMID 9249217.
- Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, et al. (April 2003). "Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene". The New England Journal of Medicine. 348 (14): 1342–1347. doi:10.1056/NEJMoa025025. PMID 12672861.
- Menichetti F (May 2005). "Current and emerging serious Gram-positive infections". Clinical Microbiology and Infection. 11 (Suppl 3): 22–28. doi:10.1111/j.1469-0691.2005.01138.x. PMID 15811021.
- Sengupta M, Jain V, Wilkinson BJ, Jayaswal RK (June 2012). "Chromatin immunoprecipitation identifies genes under direct VraSR regulation in Staphylococcus aureus". Canadian Journal of Microbiology. 58 (6): 703–708. doi:10.1139/w2012-043. PMID 22571705.
- Holasová, Kateřina; Křížkovská, Bára; Hoang, Lan; Dobiasová, Simona; Lipov, Jan; Macek, Tomáš; Křen, Vladimír; Valentová, Kateřina; Ruml, Tomáš; Viktorová, Jitka (1 May 2022). "Flavonolignans from silymarin modulate antibiotic resistance and virulence in Staphylococcus aureus". Biomedicine & Pharmacotherapy. 149: 112806. doi:10.1016/j.biopha.2022.112806. ISSN 0753-3322. PMID 35303568.
- "General Information: Community acquired MRSA". CDC. 25 March 2016.
- Latha T, Anil Bhat, Manjunatha Hande, Chiranjay Mukhopadyay, Elsa Sanatombi Devi, Baby Nayak. Methicillin-resistant staphylococcus aureus carriage among health-care professionals of a tertiary care hospital. Asian J Pharm Clin Res. 2018;11(3):346-9.
- Mello DD, Latha T, Alva J, Barnini Banarjee VK. Methicillin Resistant Staphylococcus Aureus (MRSA) Infection: Knowledge, Preventive Practices and Colonisation among Healthcare Professionals of Surgical Units. Indian Journal of Public Health Research & Development. 2019 Dec;10(12):587-91.
- Campos A, Arias A, Betancor L, Rodríguez C, Hernández AM, López Aguado D, Sierra A (July 1998). "Study of common aerobic flora of human cerumen". The Journal of Laryngology and Otology. 112 (7): 613–616. doi:10.1017/s002221510014126x. PMID 9775288. S2CID 29362695.
- Quinn GA, Cole AM (September 2007). "Suppression of innate immunity by a nasal carriage strain of Staphylococcus aureus increases its colonization on nasal epithelium". Immunology. 122 (1): 80–89. doi:10.1111/j.1365-2567.2007.02615.x. PMC 2265977. PMID 17472720.
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL (December 2005). "The role of nasal carriage in Staphylococcus aureus infections". The Lancet. Infectious Diseases. 5 (12): 751–762. doi:10.1016/S1473-3099(05)70295-4. PMID 16310147.
- Sing A, Tuschak C, Hörmansdorfer S (March 2008). "Methicillin-resistant Staphylococcus aureus in a family and its pet cat". The New England Journal of Medicine. 358 (11): 1200–1201. doi:10.1056/NEJMc0706805. PMID 18337614.
- Munir MT, Pailhories H, Eveillard M, Irle M, Aviat F, Federighi M, Belloncle C (August 2020). "Experimental Parameters Influence the Observed Antimicrobial Response of Oak Wood (Quercus petraea)". Antibiotics. 9 (9): 535. doi:10.3390/antibiotics9090535. PMC 7558063. PMID 32847132.
- Neely AN, Maley MP (February 2000). "Survival of enterococci and staphylococci on hospital fabrics and plastic". Journal of Clinical Microbiology. 38 (2): 724–726. doi:10.1128/JCM.38.2.724-726.2000. PMC 86187. PMID 10655374.
- Cook HA, Furuya EY, Larson E, Vasquez G, Lowy FD (February 2007). "Heterosexual transmission of community-associated methicillin-resistant Staphylococcus aureus". Clinical Infectious Diseases. 44 (3): 410–413. doi:10.1086/510681. PMID 17205449.
- Shafiei Y, Razavilar V, Javadi A (2011). "Thermal Death Time of Staphylococcus Aureus (PTCC=29213) and Staphylococcus Epidermidis (PTCC=1435) in Distilled Water" (PDF). Australian Journal of Basic and Applied Sciences. 5 (11): 1551–4. Archived (PDF) from the original on 2 July 2015.
- Wu X, Su YC (August 2014). "Effects of frozen storage on survival of Staphylococcus aureus and enterotoxin production in precooked tuna meat". Journal of Food Science. 79 (8): M1554–M1559. doi:10.1111/1750-3841.12530. PMID 25039601.
- Bolton KJ, Dodd CE, Mead GC, Waites WM (1988). "Chlorine resistance of strains of Staphylococcus aureus isolated from poultry processing plants". Letters in Applied Microbiology. 6 (2): 31–34. doi:10.1111/j.1472-765X.1988.tb01208.x. S2CID 84137649.
- Mead GC, Adams BW (1986). "Chlorine resistance of Staphylococcus aureus isolated from turkeys and turkey products". Letters in Applied Microbiology. 3 (6): 131–133. doi:10.1111/j.1472-765X.1986.tb01566.x. S2CID 86676949.
- van Rijen M, Bonten M, Wenzel R, Kluytmans J, et al. (Cochrane Wounds Group) (October 2008). "Mupirocin ointment for preventing Staphylococcus aureus infections in nasal carriers". The Cochrane Database of Systematic Reviews. 2011 (4): CD006216. doi:10.1002/14651858.CD006216.pub2. PMC 8988859. PMID 18843708.
- Liu Z, Norman G, Iheozor-Ejiofor Z, Wong JK, Crosbie EJ, Wilson P, et al. (Cochrane Wounds Group) (May 2017). "Nasal decontamination for the prevention of surgical site infection in Staphylococcus aureus carriers". The Cochrane Database of Systematic Reviews. 5 (8): CD012462. doi:10.1002/14651858.CD012462.pub2. PMC 6481881. PMID 28516472.
- "Food standard agency".
- "A Shot Against MRSA?" (PDF). Resources for the Future. 20 April 2009. Archived (PDF) from the original on 8 March 2016. Retrieved 7 October 2015.
- "Strengthening the immune system as an antimicrobial strategy against Staphylococcus aureus infections" (PDF). FORMATEX RESEARCH CENTER. 11 December 2013. Archived (PDF) from the original on 22 November 2015. Retrieved 7 October 2015.
- "Intercell, Merck terminate V710 Phase II/III trial against S. aureus infection". Merck & Co. 8 June 2011. Retrieved 7 October 2015.
- "Nabi Biopharmaceuticals Completes Final PentaStaph(TM) Milestone" (Press release). GLOBE NEWSWIRE. 27 April 2011. Retrieved 7 October 2015.
- "Vaccines to prevent antibiotic‐resistant Staphylococcus aureus (MRSA)infections" (PDF). University of Chicago. Archived (PDF) from the original on 10 September 2016. Retrieved 11 May 2017.
- Clinical trial number NCT01160172 for "A Study to Evaluate the Safety, Reactogenicity and Immunogenicity of GSK Biologicals' Staphylococcal Investigational Vaccine in Healthy Adults" at ClinicalTrials.gov
- "Status of vaccine research and development of vaccines for Staphylococcus aureus" (PDF). ELSEVIER. 19 April 2016. Archived (PDF) from the original on 11 October 2016. Retrieved 10 October 2016.
- "Pfizer Begins Phase 2b Study of Its Investigational Multi-antigen Staphylococcus aureus Vaccine in Adults Undergoing Elective Spinal Fusion Surgery". Pfizer Inc. 7 July 2015. Retrieved 24 February 2016.
- Clinical trial number NCT02388165 for "Safety and Efficacy of SA4Ag Vaccine in Adults Having Elective Posterior Instrumented Lumbar Spinal Fusion Procedure (STRIVE)" at ClinicalTrials.gov
- Begier E, Seiden DJ, Patton M, Zito E, Severs J, Cooper D, et al. (February 2017). "SA4Ag, a 4-antigen Staphylococcus aureus vaccine, rapidly induces high levels of bacteria-killing antibodies". Vaccine. 35 (8): 1132–1139. doi:10.1016/j.vaccine.2017.01.024. PMID 28143674.
- Torre A, Bacconi M, Sammicheli C, Galletti B, Laera D, Fontana MR, et al. (August 2015). "Four-component Staphylococcus aureus vaccine 4C-staph enhances Fcγ receptor expression in neutrophils and monocytes and mitigates S. aureus infection in neutropenic mice". Infection and Immunity. 83 (8): 3157–3163. doi:10.1128/IAI.00258-15. PMC 4496606. PMID 26015481.
- Mayr-Harting A (August 1955). "The acquisition of penicillin resistance by Staphylococcus aureus, strain Oxford". Journal of General Microbiology. 13 (1): 9–21. doi:10.1099/00221287-13-1-9. PMID 13252206.
- Kearns AM, Ganner M, Holmes A (August 2006). "The 'Oxford Staphylococcus': a note of caution". The Journal of Antimicrobial Chemotherapy. 58 (2): 480–481. doi:10.1093/jac/dkl230. PMID 16735421.
- EUCAST (1 January 2020). Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST: version 10.0. Växjö, Sweden: European Society of Clinical Microbiology and Infectious Diseases. p. 9.
Further reading
- Loskill P, Pereira PM, Jung P, Bischoff M, Herrmann M, Pinho MG, Jacobs K (September 2014). "Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the Staphylococcus aureus cell envelope". Biophysical Journal. 107 (5): 1082–1089. Bibcode:2014BpJ...107.1082L. doi:10.1016/j.bpj.2014.07.029. PMC 4156677. PMID 25185544.
- Benson MA, Ohneck EA, Ryan C, Alonzo F, Smith H, Narechania A, et al. (August 2014). "Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element". Molecular Microbiology. 93 (4): 664–681. doi:10.1111/mmi.12682. PMC 4127135. PMID 24962815.
External links
- StopMRSANow.org — Discusses how to prevent the spread of MRSA
- TheMRSA.com — Understand what the MRSA infection is all about.
- "Staphylococcus aureus". NCBI Taxonomy Browser. 1280.
- Packham C (16 March 2015). "Successful in vivo test of breakthrough Staphylococcus aureus vaccine". Medical Press. Archived from the original on 19 September 2012. Retrieved 18 March 2015.
- Type strain of Staphylococcus aureus at BacDive – the Bacterial Diversity Metadatabase