Transglutaminase
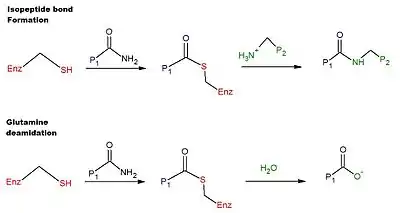
Transglutaminases are enzymes that in nature primarily catalyze the formation of an isopeptide bond between γ-carboxamide groups ( -(C=O)NH2 ) of glutamine residue side chains and the ε-amino groups ( -NH2 ) of lysine residue side chains with subsequent release of ammonia ( NH3 ). Lysine and glutamine residues must be bound to a peptide or a protein so that this cross-linking (between separate molecules) or intramolecular (within the same molecule) reaction can happen.[1] Bonds formed by transglutaminase exhibit high resistance to proteolytic degradation (proteolysis).[2] The reaction is[1]
- Gln-(C=O)NH2 + NH2-Lys → Gln-(C=O)NH-Lys + NH3
| Transglutaminase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
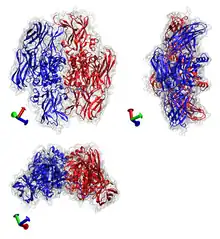 Transglutaminase example: coagulation factor XIII from human blood. PDB code: 1EVU. | |||||||||
| Identifiers | |||||||||
| EC no. | 2.3.2.13 | ||||||||
| CAS no. | 80146-85-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Transglutaminases can also join a primary amine ( RNH2 ) to the side chain carboxyamide group of a protein/peptide bound glutamine residue thus forming an isopeptide bond[1]
- Gln-(C=O)NH2 + RNH2 → Gln-(C=O)NHR + NH3
These enzymes can also deamidate glutamine residues to glutamic acid residues in the presence of water[1]
- Gln-(C=O)NH2 + H2O → Gln-COOH + NH3
Transglutaminase isolated from Streptomyces mobaraensis -bacteria for example, is a calcium-independent enzyme. Mammalian transglutaminases among other transglutaminases require Ca2+ ions as a cofactor.[1]
Transglutaminases were first described in 1959.[3] The exact biochemical activity of transglutaminases was discovered in blood coagulation protein factor XIII in 1968.[4]
Examples
Nine transglutaminases have been characterised in humans,[5] eight of which catalyse transamidation reactions. These TGases have a three or four-domain organization, with immunoglobulin-like domains surrounding the central catalytic domain. The core domain belongs to the papain-like protease superfamily (CA clan) and uses a Cys-His-Asp catalytic triad.[2] Protein 4.2 , also referred to as band 4.2, is a catalytically inactive member of the human transglutaminase family that has a Cys to Ala substitution at the catalytic triad.[6]
| Name | Gene | Activity | Chromosome | OMIM |
|---|---|---|---|---|
| Factor XIII (fibrin-stabilizing factor) chain A | F13A1 | coagulation | 6p25-p24 | 134570 |
| Keratinocyte transglutaminase | TGM1 | skin | 14q11.2 | 190195 |
| Tissue transglutaminase | TGM2 | ubiquitous | 20q11.2-q12 | 190196 |
| Epidermal transglutaminase | TGM3 | skin | 20q12 | 600238 |
| Prostate transglutaminase | TGM4 | prostate | 3p22-p21.33 | 600585 |
| TGM X | TGM5[7] | skin | 15q15.2 | 603805 |
| TGM Y | TGM6 | nerves, CNS | 20q11-15 | 613900 |
| TGM Z | TGM7 | testis, lung | 15q15.2 | 606776 |
| Protein 4.2 | EPB42 | erythrocytes, bone marrow, spleen | 15q15.2 | 177070 |
Bacterial transglutaminases are single-domain proteins with a similarly-folded core. The transglutaminase found in some bacteria runs on a Cys-Asp diad.[8]
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Biological role
Transglutaminases form extensively cross-linked, generally insoluble protein polymers. These biological polymers are indispensable for an organism to create barriers and stable structures. Examples are blood clots (coagulation factor XIII), skin, and hair. The catalytic reaction is generally viewed as being irreversible, and must be closely monitored through extensive control mechanisms.[2]
Role in disease
Deficiency of factor XIII (a rare genetic condition) predisposes to hemorrhage; concentrated enzyme can be used to correct the abnormality and reduce bleeding risk.[2]
Anti-transglutaminase antibodies are found in celiac disease and may play a role in the small bowel damage in response to dietary gliadin that characterises this condition.[2] In the related condition dermatitis herpetiformis, in which small bowel changes are often found and which responds to dietary exclusion of gliadin-containing wheat products, epidermal transglutaminase is the predominant autoantigen.[9]
Recent research indicates that sufferers from neurological diseases like Huntington's[10] and Parkinson's[11] may have unusually high levels of one type of transglutaminase, tissue transglutaminase. It is hypothesized that tissue transglutaminase may be involved in the formation of the protein aggregates that causes Huntington's disease, although it is most likely not required.[2][12]
Mutations in keratinocyte transglutaminase are implicated in lamellar ichthyosis.
Structural studies
As of late 2007, 19 structures have been solved for this class of enzymes, with PDB accession codes 1EVU, 1EX0, 1F13, 1FIE, 1G0D, 1GGT, 1GGU, 1GGY, 1IU4, 1KV3, 1L9M, 1L9N, 1NUD, 1NUF, 1NUG, 1QRK, 1RLE, 1SGX, and 1VJJ.
Industrial and culinary applications

.jpg.webp)
In commercial food processing, transglutaminase is used to bond proteins together. Examples of foods made using transglutaminase include imitation crabmeat, and fish balls. It is produced by Streptomyces mobaraensis fermentation in commercial quantities (P81453) or extracted from animal blood,[13] and is used in a variety of processes, including the production of processed meat and fish products.
Transglutaminase can be used as a binding agent to improve the texture of protein-rich foods such as surimi or ham.[14]
Thrombin–fibrinogen "meat glue" from bovine and porcine sources was banned throughout the European Union as a food additive in 2010.[15] Transglutaminase remains allowed and is not required to be declared, as it is considered a processing aid and not an additive which remains present in the final product.
Molecular gastronomy
Transglutaminase is also used in molecular gastronomy to meld new textures with existing tastes. Besides these mainstream uses, transglutaminase has been used to create some unusual foods. British chef Heston Blumenthal is credited with the introduction of transglutaminase into modern cooking.
Wylie Dufresne, chef of New York's avant-garde restaurant wd~50, was introduced to transglutaminase by Blumenthal, and invented a "pasta" made from over 95% shrimp thanks to transglutaminase.[16]
Synonyms
- protein-glutamine gamma-glutamyltransferase (systematic)
- fibrinoligase
- glutaminylpeptide gamma-glutamyltransferase
- protein-glutamine:amine gamma-glutamyltransferase
- R-glutaminyl-peptide:amine gamma-glutamyl transferase
See also
References
- DeJong GA, Koppelman SJ (2002). "Transglutaminase Catalyzed Reactions: Impact on Food Applications". Journal of Food Science. 67 (8): 2798–2806. doi:10.1111/j.1365-2621.2002.tb08819.x.
- Griffin M, Casadio R, Bergamini CM (December 2002). "Transglutaminases: nature's biological glues". The Biochemical Journal. 368 (Pt 2): 377–96. doi:10.1042/BJ20021234. PMC 1223021. PMID 12366374.
- Clarke DD, Mycek MJ, Neidle A, Waelsch H (1959). "The incorporation of amines into proteins". Arch Biochem Biophys. 79: 338–354. doi:10.1016/0003-9861(59)90413-8.
- Pisano JJ, Finlayson JS, Peyton MP (May 1968). "[Cross-link in fibrin polymerized by factor 13: epsilon-(gamma-glutamyl)lysine]". Science. 160 (3830): 892–3. Bibcode:1968Sci...160..892P. doi:10.1126/science.160.3830.892. PMID 4967475. S2CID 95459438.
- Grenard P, Bates MK, Aeschlimann D (August 2001). "Evolution of transglutaminase genes: identification of a transglutaminase gene cluster on human chromosome 15q15. Structure of the gene encoding transglutaminase X and a novel gene family member, transglutaminase Z". The Journal of Biological Chemistry. 276 (35): 33066–78. doi:10.1074/jbc.M102553200. PMID 11390390.
- Eckert RL, Kaartinen MT, Nurminskaya M, Belkin AM, Colak G, Johnson GV, Mehta K (April 2014). "Transglutaminase regulation of cell function". Physiological Reviews. 94 (2): 383–417. doi:10.1152/physrev.00019.2013. PMC 4044299. PMID 24692352.
- Aeschlimann D, Koeller MK, Allen-Hoffmann BL, Mosher DF (February 1998). "Isolation of a cDNA encoding a novel member of the transglutaminase gene family from human keratinocytes. Detection and identification of transglutaminase gene products based on reverse transcription-polymerase chain reaction with degenerate primers". The Journal of Biological Chemistry. 273 (6): 3452–60. doi:10.1074/jbc.273.6.3452. PMID 9452468.
- Kashiwagi T, Yokoyama K, Ishikawa K, Ono K, Ejima D, Matsui H, Suzuki E (November 2002). "Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense". The Journal of Biological Chemistry. 277 (46): 44252–60. doi:10.1074/jbc.M203933200. PMID 12221081.
- Sárdy M, Kárpáti S, Merkl B, Paulsson M, Smyth N (March 2002). "Epidermal transglutaminase (TGase 3) is the autoantigen of dermatitis herpetiformis". The Journal of Experimental Medicine. 195 (6): 747–57. doi:10.1084/jem.20011299. PMC 2193738. PMID 11901200.
- Karpuj MV, Becher MW, Steinman L (January 2002). "Evidence for a role for transglutaminase in Huntington's disease and the potential therapeutic implications". Neurochemistry International. 40 (1): 31–6. doi:10.1016/S0197-0186(01)00060-2. PMID 11738470. S2CID 40198925.
- Vermes I, Steur EN, Jirikowski GF, Haanen C (October 2004). "Elevated concentration of cerebrospinal fluid tissue transglutaminase in Parkinson's disease indicating apoptosis". Movement Disorders. 19 (10): 1252–4. doi:10.1002/mds.20197. PMID 15368613. S2CID 102503.
- Lesort M, Chun W, Tucholski J, Johnson GV (January 2002). "Does tissue transglutaminase play a role in Huntington's disease?". Neurochemistry International. 40 (1): 37–52. doi:10.1016/S0197-0186(01)00059-6. PMID 11738471. S2CID 7983848.
- Köhler W (22 August 2008). "Gelijmde slavink" (in Dutch). NRC Handelsblad. Archived from the original on 20 February 2009. Retrieved 5 March 2009.
- Yokoyama K, Nio N, Kikuchi Y (May 2004). "Properties and applications of microbial transglutaminase". Applied Microbiology and Biotechnology. 64 (4): 447–54. doi:10.1007/s00253-003-1539-5. PMID 14740191. S2CID 19068193.
- "EU Bans 'Meat Glue' - Food Safety News". foodsafetynews.com. 24 May 2010. Archived from the original on 5 April 2018. Retrieved 6 May 2018.
- Jon B (11 February 2005). "Noodles, reinvented". NBC News. Retrieved 2 April 2008.
Further reading
- Akbari, et al. (2021). "Recent advances in microbial transglutaminase biosynthesis and its application in the food industry". Trends in Food Science & Technology. Retrieved 31 August 2023.
- Fesus L, Hitomi K, Kojima S (2015). Transglutaminases: Multiple Functional Modifiers and Targets for New Drug Discovery. Springer Japan. ISBN 978-4-431-55823-1.
- Nuijens T, Schmidt M (2019). Enzyme-mediated ligation methods. Humana, New York, NY. ISBN 978-1-4939-9545-5.
- Kuddus M (2018). Enzymes in food technology : improvements and innovations. Springer, Singapore. ISBN 978-981-13-1932-7.
- Kelleher JB (11 May 2012). "Industry defends ingredient critics deride as "meat glue"". Chicago Tribune. Retrieved 20 July 2012.
- Marhons, et al. (2023). "Properties of yoghurt treated with microbial transglutaminase and exopolysaccharides". International Dairy Journal. Retrieved 31 August 2023.
- U.S. Patent 5,156,956 – A transglutaminase catalyzing an acyl transfer reaction of a Γ-carboxyamide group of a glutamine residue in a peptide or protein chain in the absence of Ca2+