Bacterial secretion system
Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors (mainly of proteins) to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.[2]
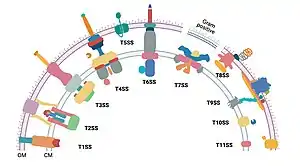
These major differences can be distinguished between Gram-negative diderm bacteria and Gram-positive monoderm bacteria. But the classification is by no means clear and complete. There are at least eight types specific to Gram-negative bacteria, four to Gram-positive bacteria, while two are common to both.[3] In addition, there is appreciable difference between diderm bacteria with lipopolysaccharide on the outer membrane (diderm-LPS) and those with mycolic acid (diderm-mycolate).[4]
Export pathways
The export pathway is responsible for crossing the inner cell membrane in diderms, and the only cell membrane in monoderms.[4]
Sec system
The general secretion (Sec) involves secretion of unfolded proteins that first remain inside the cells. In Gram-negative bacteria, the secreted protein is sent to either the inner membrane or the periplasm. But in Gram-positive bacteria, the protein can stay in the cell or is mostly transported out of the bacteria using other secretion systems. Among Gram-negative bacteria, Escherichia coli, Vibrio cholerae, Klebsiella pneumoniae, and Yersinia enterocolitica use the Sec system. Staphylococcus aureus and Listeria monocytogenes are Gram-positive bacteria that use the Sec system.[5]
The Sec system utilises two different pathways for secretion: the SecA and signal recognition particle (SRP) pathways. SecA is an ATPase motor protein and has many related proteins including SecD, SecE, SecF, SegG, SecM, and SecY. SRP is a ribonucleoprotein (protein-RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and to the cell membrane in prokaryotes. The two pathways require different molecular chaperones and ultimately use a protein-transporting channel SecYEG for transporting the proteins across the inner cell membrane.[6] In the SecA pathway, SecB acts as a chaperone, helping protein transport to the periplasm after complete synthesis of the peptide chains. Whereas in the SRP pathway, YidC is the chaperone, and transport proteins to the cell membrane while they are still undergoing peptide synthesis.[7] In Escherichia coli, inner membrane proteins are mainly targeted by the SRP pathway and outer membrane or periplasmic proteins are targeted by the SecA pathway.[8] However, a recent selective ribosome profiling study suggest that inner membrane proteins with large periplasmic loops are targeted by the SecA pathway.[9]
SecA or post-translational pathway
Proteins are synthesised in ribosomes by a process of serially adding amino acids, called translation. In SecA pathway, a chaperone trigger factor (TF) first bind to the exposed N-terminal signal sequence of the peptide chain. As elongation of peptide chain continues, TF is replaced by SecB. SecB specifically maintains the peptide in an unfolded state, and aids in the binding of SecA. The complex can then bind to SecYEG, by which SecA is activated by binding with ATP. Driven by ATP energy, SecA pushes the protein through the secYEG channel. SecD/F complex also helps in the pulling of the protein from the other side of the cell membrane.[10]
In recent years, the SecA pathway has also been suggested to have a co-translational mechanism, meaning that the polypeptide would be targeted directly by SecA during its synthesis.[11]
SRP pathway
In this pathway, SRP competes with TF and binds to the N-terminal signal sequence. Proteins from inner membrane stops the process of chain elongation. The SRP then binds to a membrane receptor, FtsY. The peptide chain-SRP-FtsY complex is then transported to SecY, where peptide elongation resumes.[7]
Tat system
The twin arginine translocation (Tat) system is similar to Sec in the process of protein secretion, however, it sends proteins only in their folded (tertiary) state. It is used by all types of bacteria, as well as archaea, and chloroplasts and mitochondria of plants.[12] In bacteria, the Tat system exports proteins from the cytoplasm across the inner cell membrane; whereas in chloroplasts, it is present in the thylakoid membrane where it aids the import of proteins from the stroma.[13] Tat proteins are highly variable in different bacteria and are classified into three major types, namely TatA, TatB, and TatC. For example, while there are only two functional Tat proteins in Bacillus subtilis,[14] there can be over a hundred in Streptomyces coelicolor.[15] Signal peptides that can recognise the Tat proteins are characterised by a consensus motif Ser/Thr-Arg-Arg-X-Phe-Leu-Lys (where X can be any polar amino acid). It is the two successive arginines from which the name twin arginine translocation came from. Replacement of any of the arginine leads to slow down or failure of secretion.[16]
Wss/Esx pathway
The Wss/Esx (ESAT-6 system) pathway is sometimes called a type VII secretion system (T7SS) despite being an export pathway.[4] It is present in Gram-positive bacteria (as WSS) and Mycobacteria (as Esx in all diderm-mycolates) such as M. tuberculosis, M. bovis, Streptomyces coelicolor and S. aureus. It is also called T7b system in Bacillus subtilis and S. aureus. It is composed of two basic components: a membrane-bound hexameric ATPase that is member of the FtsK/SpoIIIE protein family,[17] and any one of the EsxA/EsxB-related protein such as EsaA, EsaD, EsxB, EsxD, as well as Ess system (EssA, EssB, and EsxC found in S. aureus).[18] EsxA and EsxB belong to a superfamily of WXG100 proteins that form dimeric helical hairpins.
In S. aureus, T7SS secretes a large toxin called EsaD, which is a member of nuclease enzymes. EsaD is made harmless (detoxified) during its biosynthesis with the help of its counterpart antitoxin EsaG. The EsaD-EsaG complex then binds with EsaE. The EsaE portion binds to EssC, which is an enzyme ATPase of the T7SS complex. During secretion, EsaG is left in the cytoplasm, and only EsaD and EsaE are secreted together. But in some strains of S. aureus, EsaD is not produced, but two copies of EsaG-like proteins are formed instead. This might explain the occurrence of T7SS in non-pathogenic species such as B. subtilis and S. coelicolor.[19]
Secretion systems
The secretion systems are responsible for crossing the outer cell membrane or both membranes in diderms. The current nomenclature applies to diderm-LPS only, as nothing is known about what diderm-mycolate bacteria use to cross their outer membrane.[4]
Type I

Type I secretion system (T1SS or TOSS) is found in Gram-negative bacteria. It depends on chaperone activity using Hly and Tol proteins. The system activates as a signal sequence HlyA binds HlyB on the cell membrane. This signal sequence is an ABC transporter. The HlyAB complex activates HlyD which uncoils and moves to the outer cell membrane. The terminal signal is recognised by TolC in the inner membrane. The HlyA is secreted out of the outer membrane through a tunnel-like protein channel.
T1SS transports various molecules including ions, carbohydrates, drugs, proteins. The secreted molecules vary in size from the small Escherichia coli peptide colicin V, which is 10 kDa, to the Pseudomonas fluorescens cell adhesion protein LapA, which is 520 kDa.[20] Among the most well known molecules are RTX toxins and lipase enzymes.
Type II
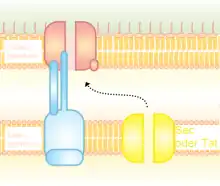
Type II (T2SS) secretion system depends on the Sec or Tat system for initial secretion inside the bacterial cell. From the periplasm, proteins are secreted out of the outer membrane secretins. Secretins are multimeric (12–14 subunits) complex of pore-forming proteins. Secretin is supported by 10–15 other inner and outer membrane proteins to constitute the complete secretion apparatus.[21]
Type III

Type III secretion system (T3SS or TTSS) is structurally similar and related to the basal body of bacterial flagella. Seen in some of the most virulent Gram-negative bacteria such as Salmonella, Shigella, Yersinia, Vibrio, it is used to inject toxic proteins into eukaryotic cells. The structure of T3SS is often described as an injectisome or needle/syringe-like apparatus. Discovered in Yersinia pestis, it was found that T3SS can inject toxins directly from the bacterial cytoplasm into the cytoplasm of its host's cells.[22]
Type IV

Type IV secretion system (T4SS or TFSS) is related to bacterial conjugation system, by which different bacteria can exchange their DNAs. The participating bacteria can be of the same or different Gram-negative bacterial species. It can transport single proteins, as well as protein-protein and DNA-protein complexes. Secretion is transferred directly from the recipient cell through the cell membranes. Agrobacterium tumefaciens, from which it was originally discovered, uses this system to send the T-DNA portion of the Ti plasmid into plant cells, in which a crown gall (tumor) is produced as a result. Helicobacter pylori uses it for delivering CagA into gastric epithelial cells, to induce gastric cancer.[23] Bordetella pertussis, the causative bacterium of whooping cough, secretes its pertussis toxin partly through T4SS. Legionella pneumophila that causes legionellosis (Legionnaires' disease) has a T4SS called icm/dot (intracellular multiplication/defect in organelle trafficking genes) that transport many bacterial proteins into its eukaryotic host.[24] More recently, it has been shown that the phytopathogen Xanthomonas citri utilizes its T4SS to secrete effectors that are lethal to other bacterial species, thus placing this system as a major fitness determinant of interspecies bacterial competition.[25][26] The prototypic Type IVA secretion system is the VirB complex of Agrobacterium tumefaciens.[27]
Type V

Type V secretion systems (T5SS) are different from other secretion systems in that they secrete themselves and only involves the outer cell membrane. For secreted protein to pass through the inner cell membrane, T5SS depends on Sec system. They have a β-barrel domain, which inserts into the outer cell membrane and forms a channel that can transport secreted protein along with it. For this activity, they are also called the autotransporter systems.[28] When the secreted proteins are exposed outside, the autotransporters are cut off (cleaved), releasing the protein from the β-barrel domain. An example of autotransporter is the Trimeric Autotransporter Adhesins.[29]
Type VI
Type VI secretion systems (T6SS) were discovered by the team of John Mekalanos at the Harvard Medical School in 2006 from Vibrio cholerae and Pseudomonas aeruginosa.[30][31] They were recognised when mutations in the Vibrio Cholerae Hcp and VrgG genes caused diminished virulence and pathogenicity.[32][33] In addition to their classic role as the pathogenicity factor, T6SS are also involved in defense against simple eukaryotic predators and in inter-bacteria interactions.[34][35] The gene for T6SS form a gene cluster that consists of more than 15 genes. Hcp and VgrG genes are the most universal genes. Structural similarity of T6SS with the tail spike of the T4 phage suggest that the process of infection is similar to that of the phage.[36]
Type VII
The T7SS of diderm-LPS bacteria is the chaperone-usher pathway (CUP).[4]
Type VIII
The T8SS of diderm-LPS bacteria is the extracellular nucleation-precipitation (ENP) pathway.[4]
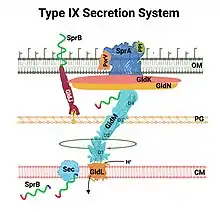
Type IX
Type IX secretion systems (T9SS) are found regularly in the Fibrobacteres-Chlorobi-Bacteroidetes lineage of bacteria, where member species include an outer membrane. The system is involved variably in one type of gliding motility, in the proper targeting of certain virulence factors to the cell surface, and the degradation of complex of biopolymers.[38] T9SS has also been known as Por (porphyrin accumulation on the cell surface) secretion,[4] after the oral pathogen Porphyromonas gingivalis. At least sixteen structural components of the system have been described, including PorU, a protein-sorting transpeptidase that removes the C-terminal sorting signal from cargo proteins and mediates their attachment instead to lipopolysaccharide.
References
- Trivedi A, Gosai J, Nakane D, Shrivastava A (2022-05-10). "Design Principles of the Rotary Type 9 Secretion System". Frontiers in Microbiology. 13: 845563. doi:10.3389/fmicb.2022.845563. PMC 9127263. PMID 35620107.
- Bocian-Ostrzycka KM, Grzeszczuk MJ, Banaś AM, Jagusztyn-Krynicka EK (May 2017). "Bacterial thiol oxidoreductases - from basic research to new antibacterial strategies". Applied Microbiology and Biotechnology. 101 (10): 3977–3989. doi:10.1007/s00253-017-8291-8. PMC 5403849. PMID 28409380.
- Green ER, Mecsas J (February 2016). Kudva IT (ed.). "Bacterial Secretion Systems: An Overview". Microbiology Spectrum (5 ed.). American Society for Microbiology Press. 4 (1): 215–239. doi:10.1128/microbiolspec.VMBF-0012-2015. ISBN 9781555819286. PMC 4804464. PMID 26999395.
- Chagnot C, Zorgani MA, Astruc T, Desvaux M (October 2013). "Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective". Frontiers in Microbiology. 4: 303. doi:10.3389/fmicb.2013.00303. PMC 3796261. PMID 24133488.
- Bensing BA, Seepersaud R, Yen YT, Sullam PM (August 2014). "Selective transport by SecA2: an expanding family of customized motor proteins". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1843 (8): 1674–1686. doi:10.1016/j.bbamcr.2013.10.019. PMC 4007388. PMID 24184206.
- Crane JM, Randall LL (November 2017). "The Sec System: Protein Export in Escherichia coli". EcoSal Plus. 7 (2): ESP–0002–2017. doi:10.1128/ecosalplus.ESP-0002-2017. PMC 5807066. PMID 29165233.
- Zhu L, Kaback HR, Dalbey RE (September 2013). "YidC protein, a molecular chaperone for LacY protein folding via the SecYEG protein machinery". The Journal of Biological Chemistry. 288 (39): 28180–28194. doi:10.1074/jbc.M113.491613. PMC 3784728. PMID 23928306.
- Denks K, Vogt A, Sachelaru I, Petriman NA, Kudva R, Koch HG (March 2014). "The Sec translocon mediated protein transport in prokaryotes and eukaryotes". Molecular Membrane Biology. 31 (2–3): 58–84. doi:10.3109/09687688.2014.907455. PMID 24762201.
- Zhu Z, Wang S, Shan SO (June 2022). "Ribosome profiling reveals multiple roles of SecA in cotranslational protein export". Nature Communications. 13 (1): 3393. doi:10.1038/s41467-022-31061-5. PMC 9192764. PMID 35697696.
- Lycklama A, Nijeholt JA, Driessen AJ (April 2012). "The bacterial Sec-translocase: structure and mechanism". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 367 (1592): 1016–1028. doi:10.1098/rstb.2011.0201. PMC 3297432. PMID 22411975.
- Huber D, Jamshad M, Hanmer R, Schibich D, Döring K, Marcomini I, et al. (January 2017). Silhavy TJ (ed.). "SecA Cotranslationally Interacts with Nascent Substrate Proteins In Vivo". Journal of Bacteriology. 199 (2). doi:10.1128/JB.00622-16. PMC 5198489. PMID 27795329.
- Yen MR, Tseng YH, Nguyen EH, Wu LF, Saier MH (June 2002). "Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system". Archives of Microbiology. 177 (6): 441–450. doi:10.1007/s00203-002-0408-4. PMID 12029389. S2CID 25129008.
- Lee PA, Tullman-Ercek D, Georgiou G (2006). "The bacterial twin-arginine translocation pathway". Annual Review of Microbiology. 60: 373–395. doi:10.1146/annurev.micro.60.080805.142212. PMC 2654714. PMID 16756481.
- Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM (December 2004). "Two minimal Tat translocases in Bacillus". Molecular Microbiology. 54 (5): 1319–1325. doi:10.1111/j.1365-2958.2004.04341.x. PMID 15554971.
- Li H, Jacques PÉ, Ghinet MG, Brzezinski R, Morosoli R (July 2005). "Determining the functionality of putative Tat-dependent signal peptides in Streptomyces coelicolor A3(2) by using two different reporter proteins". Microbiology. 151 (Pt 7): 2189–2198. doi:10.1099/mic.0.27893-0. PMID 16000709.
- Stanley NR, Palmer T, Berks BC (April 2000). "The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli". The Journal of Biological Chemistry. 275 (16): 11591–11596. doi:10.1074/jbc.275.16.11591. PMID 10766774.
- Aly KA, Anderson M, Ohr RJ, Missiakas D (December 2017). "Isolation of a Membrane Protein Complex for Type VII Secretion in Staphylococcus aureus". Journal of Bacteriology. 199 (23): e00482–17. doi:10.1128/JB.00482-17. PMC 5686593. PMID 28874412.
- Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jäger F, Cargill JS, et al. (September 2014). "Heterogeneity in ess transcriptional organization and variable contribution of the Ess/Type VII protein secretion system to virulence across closely related Staphylocccus aureus strains". Molecular Microbiology. 93 (5): 928–943. doi:10.1111/mmi.12707. PMC 4285178. PMID 25040609.
- Cao Z, Casabona MG, Kneuper H, Chalmers JD, Palmer T (October 2016). "The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria". Nature Microbiology. 2: 16183. doi:10.1038/nmicrobiol.2016.183. PMC 5325307. PMID 27723728.
- Boyd CD, Smith TJ, El-Kirat-Chatel S, Newell PD, Dufrêne YF, O'Toole GA (August 2014). "Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization". Journal of Bacteriology. 196 (15): 2775–2788. doi:10.1128/JB.01629-14. PMC 4135675. PMID 24837291.
- Korotkov KV, Sandkvist M, Hol WG (April 2012). "The type II secretion system: biogenesis, molecular architecture and mechanism". Nature Reviews. Microbiology. 10 (5): 336–351. doi:10.1038/nrmicro2762. PMC 3705712. PMID 22466878.
- Büttner D (June 2012). "Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria". Microbiology and Molecular Biology Reviews. 76 (2): 262–310. doi:10.1128/MMBR.05017-11. PMC 3372255. PMID 22688814.
- Hatakeyama M, Higashi H (December 2005). "Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis". Cancer Science. 96 (12): 835–843. doi:10.1111/j.1349-7006.2005.00130.x. PMID 16367902. S2CID 5721063.
- Cascales E, Christie PJ (November 2003). "The versatile bacterial type IV secretion systems". Nature Reviews. Microbiology. 1 (2): 137–149. doi:10.1038/nrmicro753. PMC 3873781. PMID 15035043.
- Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, et al. (March 2015). "Bacterial killing via a type IV secretion system". Nature Communications. 6: 6453. Bibcode:2015NatCo...6.6453S. doi:10.1038/ncomms7453. PMID 25743609.
- Sgro GG, Costa TR, Cenens W, Souza DP, Cassago A, Coutinho de Oliveira L, et al. (December 2018). "Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri". Nature Microbiology. 3 (12): 1429–1440. doi:10.1038/s41564-018-0262-z. PMC 6264810. PMID 30349081.
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E (2005). "Biogenesis, architecture, and function of bacterial type IV secretion systems". Annual Review of Microbiology. 59: 451–485. doi:10.1146/annurev.micro.58.030603.123630. PMC 3872966. PMID 16153176.
- Thanassi DG, Stathopoulos C, Karkal A, Li H (2005). "Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of gram-negative bacteria (review)". Molecular Membrane Biology. 22 (1–2): 63–72. doi:10.1080/09687860500063290. PMID 16092525. S2CID 2708575.
- Gerlach RG, Hensel M (October 2007). "Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens". International Journal of Medical Microbiology. 297 (6): 401–415. doi:10.1016/j.ijmm.2007.03.017. PMID 17482513.
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, et al. (January 2006). "Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system". Proceedings of the National Academy of Sciences of the United States of America. 103 (5): 1528–1533. Bibcode:2006PNAS..103.1528P. doi:10.1073/pnas.0510322103. JSTOR 30048406. PMC 1345711. PMID 16432199.
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. (June 2006). "A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus". Science. 312 (5779): 1526–1530. Bibcode:2006Sci...312.1526M. doi:10.1126/science.1128393. PMC 2800167. PMID 16763151.
- Bingle LE, Bailey CM, Pallen MJ (February 2008). "Type VI secretion: a beginner's guide" (PDF). Current Opinion in Microbiology. 11 (1): 3–8. doi:10.1016/j.mib.2008.01.006. PMID 18289922.
- Cascales E (August 2008). "The type VI secretion toolkit". EMBO Reports. 9 (8): 735–741. doi:10.1038/embor.2008.131. PMC 2515208. PMID 18617888.
- Schwarz S, Hood RD, Mougous JD (December 2010). "What is type VI secretion doing in all those bugs?". Trends in Microbiology. 18 (12): 531–537. doi:10.1016/j.tim.2010.09.001. PMC 2991376. PMID 20961764.
- Coulthurst SJ (2013). "The Type VI secretion system - a widespread and versatile cell targeting system". Research in Microbiology. 164 (6): 640–654. doi:10.1016/j.resmic.2013.03.017. PMID 23542428.
- Silverman JM, Brunet YR, Cascales E, Mougous JD (2012). "Structure and regulation of the type VI secretion system". Annual Review of Microbiology. 66: 453–472. doi:10.1146/annurev-micro-121809-151619. PMC 3595004. PMID 22746332.
- Trivedi A, Gosai J, Nakane D, Shrivastava A (2022). "Design Principles of the Rotary Type 9 Secretion System". Frontiers in Microbiology. 13: 845563. doi:10.3389/fmicb.2022.845563. PMC 9127263. PMID 35620107.
- Veith PD, Glew MD, Gorasia DG, Reynolds EC (October 2017). "Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers". Molecular Microbiology. 106 (1): 35–53. doi:10.1111/mmi.13752. hdl:11343/208056. PMID 28714554. S2CID 19387266.