Ecdysone receptor
The ecdysone receptor is a nuclear receptor found in arthropods, where it controls development and contributes to other processes such as reproduction. The receptor is a non-covalent heterodimer of two proteins, the EcR protein and ultraspiracle protein (USP). It binds to and is activated by ecdysteroids. Insect ecdysone receptors are currently better characterized than those from other arthropods, and mimics of ecdysteroids are used commercially as caterpillar-selective insecticides.
| Ecdysone receptor protein | |||||||
|---|---|---|---|---|---|---|---|
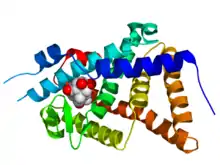 Crystallographic structure of the ligand binding domain of the ecdysone receptor (rainbow color, N-terminus = blue, C-terminus = red) from Heliothis virescens complexed with ponasterone A (space-filling model, carbon = white, oxygen = red).[1] | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | EcR | ||||||
| Alt. symbols | EcRH, NR1H1 | ||||||
| Entrez | 35540 | ||||||
| PDB | 1R0O More structures | ||||||
| RefSeq (mRNA) | NM_165461 | ||||||
| RefSeq (Prot) | NP_724456 | ||||||
| UniProt | P34021 | ||||||
| Other data | |||||||
| Chromosome | 2R: 1.97 - 2.07 Mb | ||||||
| |||||||
| Ultraspiracle protein | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | USP | ||||||
| Alt. symbols | Cf1, NR2B4 | ||||||
| Entrez | 31165 | ||||||
| PDB | 1HG4 More structures | ||||||
| RefSeq (mRNA) | NM_057433 | ||||||
| RefSeq (Prot) | NP_476781 | ||||||
| UniProt | P20153 | ||||||
| Other data | |||||||
| Chromosome | X: 1.93 - 1.94 Mb | ||||||
| |||||||
Function
Pulses of 20-hydroxyecdysone occur during insect development, whereupon this hormone binds to the ecdysone receptor, a ligand-activated transcription factor found in the nuclei of insect cells.[3] This in turn leads to the activation of many other genes, as evidenced by puffing of polytene chromosomes at over a hundred sites. Ultimately the activation cascade causes physiological changes that result in ecdysis (moulting). The temporal expression of ecdysone receptor within neural stem cells mediates temporal patterning and neural diversity.[4][5]
Structure
The receptor is a non-covalent heterodimer of two proteins, the EcR protein and ultraspiracle protein (USP). These nuclear hormone receptor proteins are the insect orthologs of the mammalian farnesoid X receptor (FXR) and retinoid X receptor (RXR) proteins, respectively. Based on sequence homology considerations,[6] some researchers reserve the term USP for the EcR partner from lepidopteran and dipteran insects, and use RXR in all other instances.
EcR and USP share the multi-domain architecture common to all nuclear hormone receptors, namely an N-terminal transcriptional activation domain (A/B domain), a DNA-binding domain (C domain, highly conserved between receptors), a linker region (D region), a ligand-binding domain (E domain, moderately conserved), and in some cases a distinct C-terminal extension (F-domain).[7] The DNA-binding domains of EcR and USP recognise specific short sequences in DNA, and mediate the binding of the heterodimer to these ecdysone response elements (ECREs) in the promoters of ecdysone-responsive genes.
The ecdysteroid-binding pocket is located in the ligand binding domain of the EcR subunit, but EcR must be dimerised with a USP (or with an RXR) for high-affinity ligand binding to occur. In such circumstances, the binding of an agonist ligand triggers a conformational change in the C-terminal part of the EcR ligand-binding domain that leads to transcriptional activation of genes under ECRE control.[8] There is also a ligand-binding pocket in the corresponding domain of USP. Its natural ligand remains uncertain, and USPs appear to be locked permanently in an inactive conformation.[9]
X-ray crystal structures have been determined for several heterodimeric DNA-binding domains[10] and ligand-binding domains[1][11] from ecdysone receptors.
Commercial applications
Ecdysone receptors have two main fields of application:
- Gene switches - ecdysone receptor-controlled transgenes for controlled gene expression in scientific research, agriculture, and medicine.[12] The last category includes potential use in gene therapy.[13]
- Insecticides - the development of selective insect growth regulators for use as environmentally benign insecticides. Although dibenzoylhydrazine compounds are not ecdysteroids, many are agonists of lepidopteran ecdysone receptors and are used as caterpillar-selective larvicides.[14][15]
References
- PDB: 1R1K; Billas IM, Iwema T, Garnier JM, Mitschler A, Rochel N, Moras D (November 2003). "Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor". Nature. 426 (6962): 91–6. Bibcode:2003Natur.426...91B. doi:10.1038/nature02112. PMID 14595375. S2CID 4413300.
- Clayton GM, Peak-Chew SY, Evans RM, Schwabe JW (February 2001). "The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation". Proceedings of the National Academy of Sciences of the United States of America. 98 (4): 1549–54. doi:10.1073/pnas.041611298. PMC 29294. PMID 11171988.
- Riddiford LM, Cherbas P, Truman JW (2000). "Ecdysone receptors and their biological actions". Vitam. Horm. Vitamins & Hormones. 60: 1–73. doi:10.1016/S0083-6729(00)60016-X. ISBN 978-0-12-709860-9. PMID 11037621.
- Syed MH, Mark B, Doe CQ (April 2017). "Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity". eLife. 6: e26287. doi:10.7554/eLife.26287. PMC 5403213. PMID 28394252.
- Henrich VC (2005). "The ecdysteroid receptor". In Gill SS, Gilbert LI, Iatrou K (eds.). Comprehensive molecular insect science. Amsterdam: Elsevier. pp. 243–285. ISBN 978-0-444-51516-2.
- Hayward DC, Bastiani MJ, Trueman JW, Truman JW, Riddiford LM, Ball EE (September 1999). "The sequence of Locusta RXR, homologous to Drosophila Ultraspiracle, and its evolutionary implications". Dev. Genes Evol. 209 (9): 564–71. doi:10.1007/s004270050290. PMID 10502114. S2CID 8703952.
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS (October 1991). "The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily". Cell. 67 (1): 59–77. doi:10.1016/0092-8674(91)90572-G. PMID 1913820. S2CID 6805386.
- Bourguet W, Germain P, Gronemeyer H (October 2000). "Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications". Trends Pharmacol. Sci. 21 (10): 381–8. doi:10.1016/S0165-6147(00)01548-0. PMID 11050318.
- Clayton GM, Peak-Chew SY, Evans RM, Schwabe JW (February 2001). "The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation". Proc. Natl. Acad. Sci. U.S.A. 98 (4): 1549–54. doi:10.1073/pnas.041611298. PMC 29294. PMID 11171988.
- Devarakonda S, Harp JM, Kim Y, Ozyhar A, Rastinejad F (November 2003). "Structure of the heterodimeric ecdysone receptor DNA-binding complex". EMBO J. 22 (21): 5827–40. doi:10.1093/emboj/cdg569. PMC 275426. PMID 14592980.
- Carmichael JA, Lawrence MC, Graham LD, Pilling PA, Epa VC, Noyce L, Lovrecz G, Winkler DA, Pawlak-Skrzecz A, Eaton RE, Hannan GN, Hill RJ (June 2005). "The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design". J. Biol. Chem. 280 (23): 22258–69. doi:10.1074/jbc.M500661200. PMID 15809296.
- Palli SR, Hormann RE, Schlattner U, Lezzi M (2005). "Ecdysteroid receptors and their applications in agriculture and medicine". Vitam. Horm. Vitamins & Hormones. 73: 59–100. doi:10.1016/S0083-6729(05)73003-X. ISBN 0-12-709873-9. PMID 16399408.
- Graham LD (June 2002). "Ecdysone-controlled expression of transgenes". Expert Opin Biol Ther. 2 (5): 525–35. doi:10.1517/14712598.2.5.525. PMID 12079488. S2CID 32235742.
- Dhadialla TS, Carlson GR, Le DP (1998). "New insecticides with ecdysteroidal and juvenile hormone activity". Annu. Rev. Entomol. 43: 545–69. doi:10.1146/annurev.ento.43.1.545. PMID 9444757.
- Dhadialla TS, Retnakaran A, Smagghe G (2005). "Insect growth- and development-disrupting insecticides". In Sarjeet S. Gill, Gilbert, Lawrence I., Kostas Iatrou (eds.). Comprehensive molecular insect science. Amsterdam: Elsevier. pp. 55–115. ISBN 0-444-51516-X.
External links
- PDB representative structures of ecdysone receptor ligand binding domains:
- 1R1K, ligand-binding domain heterodimer of Heliothis viresecens in complex with an ecdysteroid; 1R20, the same heterodimer in complex with a dibenzoylhydrazine agonist.
- Mimic insecticide, and associated regulatory information (for Australia).