Urease
Ureases (EC 3.5.1.5), functionally, belong to the superfamily of amidohydrolases and phosphotriesterases.[2] Ureases are found in numerous bacteria, fungi, algae, plants, and some invertebrates, as well as in soils, as a soil enzyme. They are nickel-containing metalloenzymes of high molecular weight.[3]
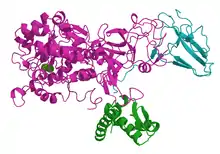 3D model of urease from Klebsiella aerogenes, two Ni2+-ions are shown as green spheres.[1] | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| EC no. | 3.5.1.5 | ||||||||
| CAS no. | 9002-13-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
These enzymes catalyze the hydrolysis of urea into carbon dioxide and ammonia:
- (NH2)2CO + H2O CO2 + 2NH3
The hydrolysis of urea occurs in two stages. In the first stage, ammonia and carbamic acid are produced. The carbamate spontaneously and rapidly hydrolyzes to ammonia and carbonic acid. Urease activity increases the pH of its environment as ammonia is produced, which is basic.
History
Its activity was first identified in 1876 by Frédéric Alphonse Musculus as a soluble ferment.[4] In 1926, James B. Sumner, showed that urease is a protein by examining its crystallized form.[5] Sumner's work was the first demonstration that a protein can function as an enzyme and led eventually to the recognition that most enzymes are in fact proteins. Urease was the first enzyme crystallized. For this work, Sumner was awarded the Nobel prize in chemistry in 1946.[6] The crystal structure of urease was first solved by P. A. Karplus in 1995.[5]
Structure
A 1984 study focusing on urease from jack bean found that the active site contains a pair of nickel centers.[7] In vitro activation also has been achieved with manganese and cobalt in place of nickel.[8] Lead salts are inhibiting.
The molecular weight is either 480 kDa or 545 kDa for jack-bean urease (calculated mass from the amino acid sequence). 840 amino acids per molecule, of which 90 are cysteine residues.[9]
The optimum pH is 7.4 and optimum temperature is 60 °C. Substrates include urea and hydroxyurea.
Bacterial ureases are composed of three distinct subunits, one large catalytic (α 60–76kDa) and two small (β 8–21 kDa, γ 6–14 kDa) commonly forming (αβγ)3 trimers stoichiometry with a 2-fold symmetric structure (note that the image above gives the structure of the asymmetric unit, one-third of the true biological assembly), they are cysteine-rich enzymes, resulting in the enzyme molar masses between 190 and 300kDa.[9]
An exceptional urease is obtained from Helicobacter sp.. These are composed of two subunits, α(26–31 kDa)-β(61–66 kDa). These subunits form a supramolecular (αβ)12 dodecameric complex.[10] of repeating α-β subunits, each coupled pair of subunits has an active site, for a total of 12 active sites.[10] It plays an essential function for survival, neutralizing gastric acid by allowing urea to enter into periplasm via a proton-gated urea channel.[11] The presence of urease is used in the diagnosis of Helicobacter species.
All bacterial ureases are solely cytoplasmic, except for those in Helicobacter pylori, which along with its cytoplasmic activity, has external activity with host cells. In contrast, all plant ureases are cytoplasmic.[9]
Fungal and plant ureases are made up of identical subunits (~90 kDa each), most commonly assembled as trimers and hexamers. For example, jack bean urease has two structural and one catalytic subunit. The α subunit contains the active site, it is composed of 840 amino acids per molecule (90 cysteines), its molecular mass without Ni(II) ions amounting to 90.77 kDa. The mass of the hexamer with the 12 nickel ions is 545.34 kDa. Other examples of homohexameric structures of plant ureases are those of soybean, pigeon pea and cotton seeds enzymes.[9]
It is important to note, that although composed of different types of subunits, ureases from different sources extending from bacteria to plants and fungi exhibit high homology of amino acid sequences. The single plant urease chain is equivalent to a fused γ-β-α organization. The Helicobacter "α" is equivalent to a fusion of the normal bacterial γ-β subunits, while its "β" subunit is equivalent to the normal bacterial α.[9] The three-chain organization is likely ancestral.[12]
Activity
The kcat/Km of urease in the processing of urea is 1014 times greater than the rate of the uncatalyzed elimination reaction of urea.[5] There are many reasons for this observation in nature. The proximity of urea to active groups in the active site along with the correct orientation of urea allow hydrolysis to occur rapidly. Urea alone is very stable due to the resonance forms it can adopt. The stability of urea is understood to be due to its resonance energy, which has been estimated at 30–40 kcal/mol.[5] This is because the zwitterionic resonance forms all donate electrons to the carbonyl carbon making it less of an electrophile making it less reactive to nucleophilic attack.[5]
Active site
The active site of ureases is located in the α (alpha) subunits. It is a bis-μ-hydroxo dimeric nickel center, with an interatomic distance of ~3.5 Å.[5] > The Ni(II) pair are weakly antiferromagnetically coupled.[13] X-ray absorption spectroscopy (XAS) studies of Canavalia ensiformis (jack bean), Klebsiella aerogenes and Sporosarcina pasteurii (formerly known as Bacillus pasteurii)[14] confirm 5–6 coordinate nickel ions with exclusively O/N ligation, including two imidazole ligands per nickel.[8] Urea substrate is proposed to displace aquo ligands.
Water molecules located towards the opening of the active site form a tetrahedral cluster that fills the cavity site through hydrogen bonds. Some amino acid residues are proposed to form mobile flap of the site, which gate for the substrate.[3] Cysteine residues are common in the flap region of the enzymes, which have been determined not to be essential in catalysis, although involved in positioning other key residues in the active site appropriately.[15] In Sporosarcina pasteurii urease, the flap was found in the open conformation, while its closed conformation is apparently needed for the reaction.[14]
When compared, the α subunits of Helicobacter pylori urease and other bacterial ureases align with the jack bean ureases.[15]
The binding of urea to the active site of urease has not been observed.[9]
Blakeley/Zerner
One mechanism for the catalysis of this reaction by urease was proposed by Blakely and Zerner.[16] It begins with a nucleophilic attack by the carbonyl oxygen of the urea molecule onto the 5-coordinate Ni (Ni-1). A weakly coordinated water ligand is displaced in its place. A lone pair of electrons from one of the nitrogen atoms on the Urea molecule creates a double bond with the central carbon, and the resulting NH2− of the coordinated substrate interacts with a nearby positively charged group. Blakeley and Zerner proposed this nearby group to be a Carboxylate ion, although deprotonated carboxylates are negatively charged.
A hydroxide ligand on the six coordinate Ni is deprotonated by a base. The carbonyl carbon is subsequently attacked by the electronegative oxygen. A pair of electrons from the nitrogen-carbon double bond returns to the nitrogen and neutralizes the charge on it, while the now 4-coordinate carbon assumes an intermediate tetrahedral orientation.
The breakdown of this intermediate is then helped by a sulfhydryl group of a cysteine located near the active site. A hydrogen bonds to one of the nitrogen atoms, breaking its bond with carbon, and releasing an NH3 molecule. Simultaneously, the bond between the oxygen and the 6-coordinate nickel is broken. This leaves a carbamate ion coordinated to the 5-coordinate Ni, which is then displaced by a water molecule, regenerating the enzyme.
The carbamate produced then spontaneously degrades to produce another ammonia and carbonic acid.[17]
Hausinger/Karplus
The mechanism proposed by Hausinger and Karplus attempts to revise some of the issues apparent in the Blakely and Zerner pathway, and focuses on the positions of the side chains making up the urea-binding pocket.[5] From the crystal structures from K. aerogenes urease, it was argued that the general base used in the Blakely mechanism, His320, was too far away from the Ni2-bound water to deprotonate in order to form the attacking hydroxide moiety. In addition, the general acidic ligand required to protonate the urea nitrogen was not identified.[18] Hausinger and Karplus suggests a reverse protonation scheme, where a protonated form of the His320 ligand plays the role of the general acid and the Ni2-bound water is already in the deprotonated state.[5] The mechanism follows the same path, with the general base omitted (as there is no more need for it) and His320 donating its proton to form the ammonia molecule, which is then released from the enzyme. While the majority of the His320 ligands and bound water will not be in their active forms (protonated and deprotonated, respectively,) it was calculated that approximately 0.3% of total urease enzyme would be active at any one time.[5] While logically, this would imply that the enzyme is not very efficient, contrary to established knowledge, usage of the reverse protonation scheme provides an advantage in increased reactivity for the active form, balancing out the disadvantage.[5] Placing the His320 ligand as an essential component in the mechanism also takes into account the mobile flap region of the enzyme. As this histidine ligand is part of the mobile flap, binding of the urea substrate for catalysis closes this flap over the active site and with the addition of the hydrogen bonding pattern to urea from other ligands in the pocket, speaks to the selectivity of the urease enzyme for urea.[5]
Ciurli/Mangani
The mechanism proposed by Ciurli and Mangani[19] is one of the more recent and currently accepted views of the mechanism of urease and is based primarily on the different roles of the two nickel ions in the active site.[14] One of which binds and activates urea, the other nickel ion binds and activates the nucleophilic water molecule.[14] With regards to this proposal, urea enters the active site cavity when the mobile ‘flap’ (which allows for the entrance of urea into the active site) is open. Stability of the binding of urea to the active site is achieved via a hydrogen-bonding network, orienting the substrate into the catalytic cavity.[14] Urea binds to the five-coordinated nickel (Ni1) with the carbonyl oxygen atom. It approaches the six-coordinated nickel (Ni2) with one of its amino groups and subsequently bridges the two nickel centers.[14] The binding of the urea carbonyl oxygen atom to Ni1 is stabilized through the protonation state of Hisα222 Nԑ. Additionally, the conformational change from the open to closed state of the mobile flap generates a rearrangement of Alaα222 carbonyl group in such a way that its oxygen atom points to Ni2.[14] The Alaα170 and Alaα366 are now oriented in a way that their carbonyl groups act as hydrogen-bond acceptors towards NH2 group of urea, thus aiding its binding to Ni2.[14] Urea is a very poor chelating ligand due to low Lewis base character of its NH2 groups. However the carbonyl oxygens of Alaα170 and Alaα366 enhance the basicity of the NH2 groups and allow for binding to Ni2.[14] Therefore, in this proposed mechanism, the positioning of urea in the active site is induced by the structural features of the active site residues which are positioned to act as hydrogen-bond donors in the vicinity of Ni1 and as acceptors in the vicinity of Ni2.[14] The main structural difference between the Ciurli/Mangani mechanism and the other two is that it incorporates a nitrogen, oxygen bridging urea that is attacked by a bridging hydroxide.[17]
Action in pathogenesis
Bacterial ureases are often the mode of pathogenesis for many medical conditions. They are associated with hepatic encephalopathy / Hepatic coma, infection stones, and peptic ulceration.[20]
Infection stones
Infection induced urinary stones are a mixture of struvite (MgNH4PO4•6H2O) and carbonate apatite [Ca10(PO4)6•CO3].[20] These polyvalent ions are soluble but become insoluble when ammonia is produced from microbial urease during urea hydrolysis, as this increases the surrounding environments pH from roughly 6.5 to 9.[20] The resultant alkalinization results in stone crystallization.[20] In humans the microbial urease, Proteus mirabilis, is the most common in infection induced urinary stones.[21]
Urease in hepatic encephalopathy / hepatic coma
Studies have shown that Helicobacter pylori along with cirrhosis of the liver cause hepatic encephalopathy and hepatic coma.[22] Helicobacter pylori release microbial ureases into the stomach. The urease hydrolyzes urea to produce ammonia and carbonic acid. As the bacteria are localized to the stomach ammonia produced is readily taken up by the circulatory system from the gastric lumen.[22] This results in elevated ammonia levels in the blood, a condition known as hyperammonemia; eradication of Helicobacter pylori show marked decreases in ammonia levels.[22]
Urease in peptic ulcers
Helicobacter pylori is also the cause of peptic ulcers with its manifestation in 55–68% reported cases.[23] This was confirmed by decreased ulcer bleeding and ulcer reoccurrence after eradication of the pathogen.[23] In the stomach there is an increase in pH of the mucosal lining as a result of urea hydrolysis, which prevents movement of hydrogen ions between gastric glands and gastric lumen.[20] In addition, the high ammonia concentrations have an effect on intercellular tight junctions increasing permeability and also disrupting the gastric mucous membrane of the stomach.[20][24]
Occurrence and applications in agriculture
Urea is found naturally in the environment and is also artificially introduced, comprising more than half of all synthetic nitrogen fertilizers used globally.[25] Heavy use of urea is thought to promote eutrophication, despite the observation that urea is rapidly transformed by microbial ureases, and thus usually does not persist.[26] Environmental urease activity is often measured as an indicator of the health of microbial communities. In the absence of plants, urease activity in soil is generally attributed to heterotrophic microorganisms, although it has been demonstrated that some chemoautotrophic ammonium oxidizing bacteria are capable of growth on urea as a sole source of carbon, nitrogen, and energy.[27]
Inhibition in fertilizers
The inhibition of urease is a significant goal in agriculture because the rapid breakdown of urea-based fertilizers is wasteful and environmentally damaging.[28] Phenyl phosphorodiamidate and N-(n-butyl)thiophosphoric triamide are two such inhibitors.[29]
Biomineralization
By promoting the formation of calcium carbonate, ureases are potentially useful for biomineralization-inspired processes.[30] Notably, micro-biologically induced formation of calcium carbonate can be used in making bioconcrete.[31]
Non-enzymatic action
In addition to acting as an enzyme, some ureases (especially plant ones) have additional effects that persist even when the catalytic function is disabled. These include entomotoxicity, inhibition of fungi, neurotoxicity in mammals, promotion of endocytosis and inflammatory eicosanoid production in mammals, and induction of chemotaxis in bacteria. These activities may be part of a defense mechanism.[12]
Urease insect-toxicity was originally noted in canatoxin, an orthologous isoform of jack bean urease. Digestion of the peptide identified a 10-kDa portion most responsible for this effect, termed jaburetox. An analogous portion from the soybean urease is named soyuretox. Studies on insects show that the entire protein is toxic without needing any digestion, however. Nevertheless, the "uretox" peptides, being more concentrated in toxicity, show promise as biopesticides.[12]
As diagnostic test
Many gastrointestinal or urinary tract pathogens produce urease, enabling the detection of urease to be used as a diagnostic to detect presence of pathogens.
Urease-positive pathogens include:
- Proteus mirabilis and Proteus vulgaris
- Ureaplasma urealyticum, a relative of Mycoplasma spp.
- Nocardia
- Corynebacterium urealyticum
- Cryptococcus spp., an opportunistic fungus
- Helicobacter pylori
- Certain Enteric bacteria including Proteus spp., Klebsiella spp., Morganella, Providencia, and possibly Serratia spp.
- Brucella
- Staphylococcus saprophyticus
- Staphylococcus aureus[32]
Ligands
Inhibitors
A wide range of urease inhibitors of different structural families are known. Inhibition of urease is not only of interest to agriculture, but also to medicine as pathogens like H. pylori produce urease as a survival mechanism. Known structural classes of inhibitors include:[33][34]
- Analogues of urea, the strongest being thioureas like 1-(4-chlorophenyl)-3-palmitoylthiourea.
- Phosphoramidates, the most commonly used in agriculture (see above).
- Hydroquinone and quinones. In medicine, the most interesting are quinolones, already a class of widely used antibiotics.
- Some plant metabolites are also urease inhibitors, an example being allicin. These have potential both as environmentally-friendly fertilizer additives[35] and natural drugs.
Extraction
First isolated as a crystal in 1926 by Sumner, using acetone solvation and centrifuging.[36] Modern biochemistry has increased its demand for urease. Jack bean meal,[37] watermelon seeds,[38] and pea seeds[39] have all proven useful sources of urease.
References
- PDB: 2KAU; Jabri E, Carr MB, Hausinger RP, Karplus PA (May 1995). "The crystal structure of urease from Klebsiella aerogenes". Science. 268 (5213): 998–1004. Bibcode:1995Sci...268..998J. doi:10.1126/science.7754395. PMID 7754395.
- Holm L, Sander C (1997). "An evolutionary treasure: unification of a broad set of amidohydrolases related to urease". Proteins. 28 (1): 72–82. CiteSeerX 10.1.1.621.2752. doi:10.1002/(SICI)1097-0134(199705)28:1<72::AID-PROT7>3.0.CO;2-L. PMID 9144792. S2CID 38845090.
- Krajewska B, van Eldik R, Brindell M (13 August 2012). "Temperature- and pressure-dependent stopped-flow kinetic studies of jack bean urease. Implications for the catalytic mechanism". Journal of Biological Inorganic Chemistry. 17 (7): 1123–1134. doi:10.1007/s00775-012-0926-8. PMC 3442171. PMID 22890689.
- Musculus, « Sur le ferment de l'urée », Comptes rendus de l'Académie des sciences, vol. 82, 1876, pp. 333-336, reachable in Gallica
- Karplus PA, Pearson MA, Hausinger RP (1997). "70 years of crystalline urease: What have we learned?". Accounts of Chemical Research. 30 (8): 330–337. doi:10.1021/ar960022j.
- The Nobel Prize in Chemistry 1946
- Anke M, Groppel B, Kronemann H, Grün M (1984). "Nickel--an essential element". IARC Sci. Publ. (53): 339–65. PMID 6398286.
- Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP (1 January 2009). "Interplay of metal ions and urease". Metallomics. 1 (3): 207–21. doi:10.1039/b903311d. PMC 2745169. PMID 20046957.
- Krajewska B (30 June 2009). "Ureases I. Functional, catalytic and kinetic properties: A review". Journal of Molecular Catalysis B: Enzymatic. 59 (1–3): 9–21. doi:10.1016/j.molcatb.2009.01.003.
- Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH (31 May 2001). "Supramolecular assembly and acid resistance of Helicobacter pylori urease". Nature Structural Biology. 8 (6): 505–509. doi:10.1038/88563. PMID 11373617. S2CID 26548257.
- Strugatsky D, McNulty R, Munson K, Chen CK, Soltis SM, Sachs G, Luecke H (8 December 2012). "Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori". Nature. 493 (7431): 255–258. doi:10.1038/nature11684. PMC 3974264. PMID 23222544.
- Kappaun, K; Piovesan, AR; Carlini, CR; Ligabue-Braun, R (September 2018). "Ureases: Historical aspects, catalytic, and non-catalytic properties - A review". Journal of Advanced Research. 13: 3–17. doi:10.1016/j.jare.2018.05.010. PMC 6077230. PMID 30094078.
- Ciurli S, Benini S, Rypniewski WR, Wilson KS, Miletti S, Mangani S (1999). "Structural properties of the nickel ions in urease: novel insights into the catalytic and inhibition mechanisms". Coordination Chemistry Reviews. 190–192: 331–355. doi:10.1016/S0010-8545(99)00093-4.
- Benini S, Rypniewski WR, Wilson KS, Miletti S, Ciurli S, Mangani S (31 January 1999). "A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels". Structure. 7 (2): 205–216. doi:10.1016/S0969-2126(99)80026-4. PMID 10368287.
- Martin PR, Hausinger RP (Oct 5, 1992). "Site-directed mutagenesis of the active site cysteine in Klebsiella aerogenes urease". The Journal of Biological Chemistry. 267 (28): 20024–7. doi:10.1016/S0021-9258(19)88659-3. PMID 1400317.
- Dixon NE, Riddles PW, Gazzola C, Blakeley RL, Zerner B (1979). "Jack Jack Bean Urease (EC3.5.1.5). V. On the Mechanism of action of urease on urea, formamide, acetamide,N-methylurea, and related compounds". Canadian Journal of Biochemistry. 58 (12): 1335–1344. doi:10.1139/o80-181. PMID 6788353.
- Zimmer M (Apr 2000). "Molecular mechanics evaluation of the proposed mechanisms for the degradation of urea by urease". J Biomol Struct Dyn. 17 (5): 787–97. doi:10.1080/07391102.2000.10506568. PMID 10798524. S2CID 41497756.
- Jabri E, Carr MB, Hausinger RP, Karplus PA (May 19, 1995). "The crystal structure of urease from Klebsiella aerogenes". Science. 268 (5213): 998–1004. Bibcode:1995Sci...268..998J. doi:10.1126/science.7754395. PMID 7754395.
- Zambelli B, Musiani F, Benini S, Ciurli S (19 July 2011). "Chemistry of Ni2+ in Urease: Sensing, Trafficking, and Catalysis". Accounts of Chemical Research. 44 (7): 520–530. doi:10.1021/ar200041k. PMID 21542631.
- Mobley HL, Hausinger RP (March 1989). "Microbial ureases: significance, regulation, and molecular characterization". Microbiological Reviews. 53 (1): 85–108. doi:10.1128/MMBR.53.1.85-108.1989. PMC 372718. PMID 2651866.
- Rosenstein IJ (1 January 1986). "Urinary Calculi: Microbiological and Crystallographic Studies". Critical Reviews in Clinical Laboratory Sciences. 23 (3): 245–277. doi:10.3109/10408368609165802. PMID 3524996.
- Agrawal A, Gupta A, Chandra M, Koowar S (17 March 2011). "Role of Helicobacter pylori infection in the pathogenesis of minimal hepatic encephalopathy and effect of its eradication". Indian Journal of Gastroenterology. 30 (1): 29–32. doi:10.1007/s12664-011-0087-7. PMID 21416318. S2CID 25452909.
- Tang JH, Liu NJ, Cheng HT, Lee CS, Chu YY, Sung KF, Lin CH, Tsou YK, Lien JM, Cheng CL (February 2009). "Endoscopic diagnosis of Helicobacter pylori infection by rapid urease test in bleeding peptic ulcers: a prospective case-control study". Journal of Clinical Gastroenterology. 43 (2): 133–9. doi:10.1097/MCG.0b013e31816466ec. PMID 19230239. S2CID 27784917.
- Caron TJ, Scott KE, Fox JG, Hagen SJ (October 2015). "Tight junction disruption: Helicobacter pylori and dysregulation of the gastric mucosal barrier". World Journal of Gastroenterology. 21 (40): 11411–27. doi:10.3748/wjg.v21.i40.11411. PMC 4616217. PMID 26523106.
- Glibert P, Harrison J, Heil C, Seitzinger S (2006). "Escalating worldwide use of urea – a global change contributing to coastal eutrophication". Biogeochemistry. 77 (3): 441–463. doi:10.1007/s10533-005-3070-5. S2CID 2209850.
- Daigh AL, Savin MC, Brye K, Norman R, Miller D (2014). "Urea persistence in floodwater and soil used for flooded rice production". Soil Use and Management. 30 (4): 463–470. doi:10.1111/sum.12142. S2CID 97961385.
- Marsh KL, Sims GK, Mulvaney RL (November 2005). "Availability of urea to autotrophic ammonia-oxidizing bacteria as related to the fate of 14 C-and 15 N-labeled urea added to soil". Biology and Fertility of Soils. 42 (2): 137–145. doi:10.1007/s00374-005-0004-2. S2CID 6245255.
- Pan B, Lam SK, Mosier A, Luo Y, Chen D (2016). "Ammonia Volatilization from Synthetic Fertilizers and its Mitigation Strategies: A Global Synthesis". Agriculture, Ecosystems & Environment. 232: 283–289. doi:10.1016/j.agee.2016.08.019.
- Gholivand K, Pooyan M, Mohammadpanah F, Pirastefar F, Junk PC, Wang J, et al. (May 2019). "Synthesis, crystal structure and biological evaluation of new phosphoramide derivatives as urease inhibitors using docking, QSAR and kinetic studies". Bioorganic Chemistry. 86: 482–493. doi:10.1016/j.bioorg.2019.01.064. PMID 30772649. S2CID 73460771.
- Anbu P, Kang CH, Shin YJ, So JS (1 March 2016). "Formations of calcium carbonate minerals by bacteria and its multiple applications". SpringerPlus. 5: 250. doi:10.1186/s40064-016-1869-2. PMC 4771655. PMID 27026942.
- Moneo S (11 September 2015). "Dutch scientist invents self-healing concrete with bacteria". Journal Of Commerce. Retrieved 23 March 2018.
- Zhou C, Bhinderwala F, Lehman MK, Thomas VC, Chaudhari SS, Yamada KJ, et al. (January 2019). "Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection". PLOS Pathogens. 15 (1): e1007538. doi:10.1371/journal.ppat.1007538. PMC 6343930. PMID 30608981.
- Modolo, LV; da-Silva, CJ; Brandão, DS; Chaves, IS (September 2018). "A minireview on what we have learned about urease inhibitors of agricultural interest since mid-2000s". Journal of Advanced Research. 13: 29–37. doi:10.1016/j.jare.2018.04.001. PMC 6077229. PMID 30094080.
- Kafarski, P; Talma, M (September 2018). "Recent advances in design of new urease inhibitors: A review". Journal of Advanced Research. 13: 101–112. doi:10.1016/j.jare.2018.01.007. PMC 6077125. PMID 30094085.
- Ee Huey, Choo; Zaireen Nisa Yahya, Wan; Mansor, Nurlidia (2019). "Allicin incorporation as urease inhibitor in a chitosan/starch based biopolymer for fertilizer application". Materials Today: Proceedings. 16: 2187–2196. doi:10.1016/j.matpr.2019.06.109. S2CID 202073615.
- Gorin G, Butler MF, Katyal JM, Buckley JE (1959). "Isolation of crystalline urease" (PDF). Proceedings of the Oklahoma Academy of Science. 40: 62–70. Retrieved Dec 7, 2014.
- Sung HY, Lee WM, Chiou MJ, Chang CT (October 1989). "A procedure for purifying jack bean urease for clinical use". Proceedings of the National Science Council, Republic of China. Part B, Life Sciences. 13 (4): 250–7. PMID 2517764.
- Prakash O, Bhushan G (January 1997). "Isolation, purification and partial characterisation of urease from seeds of water melon (Citrullus vulgaris)". Journal of Plant Biochemistry and Biotechnology. 6: 45–47. doi:10.1007/BF03263009. S2CID 41143649.
- El-Hefnawy ME, Sakran M, Ismail AI, Aboelfetoh EF (July 2014). "Extraction, purification, kinetic and thermodynamic properties of urease from germinating Pisum sativum L. seeds". BMC Biochemistry. 15 (1): 15. doi:10.1186/1471-2091-15-15. PMC 4121304. PMID 25065975.
External links
- Mobley HL (2001). "Chapter 16:Urease". In Mobley HL, Mendz GL, Hazell SL (eds.). Helicobacter pylori: Physiology and Genetics. Washington (DC): ASM Press. ISBN 978-1-55581-213-3. PMID 21290719.