G418
G418 (Geneticin) is an aminoglycoside antibiotic similar in structure to gentamicin B1. It is produced by Micromonospora rhodorangea.[1] G418 blocks polypeptide synthesis by inhibiting the elongation step in both prokaryotic and eukaryotic cells.[1] Resistance to G418 is conferred by the neo gene from Tn5 encoding an aminoglycoside 3'-phosphotransferase, APT 3' II.[1] G418 is an analog of neomycin sulfate, and has similar mechanism as neomycin. G418 is commonly used in laboratory research to select genetically engineered cells .[2] In general for bacteria and algae concentrations of 5 μg/mL or less are used, for mammalian cells concentrations of approximately 400 μg/mL are used for selection and 200 μg/mL for maintenance. However, optimal concentration for resistant clones selection in mammalian cells depends on the cell line used as well as on the plasmid carrying the resistance gene, therefore antibiotic titration should be done to find the best condition for every experimental system. Titration should be done using antibiotic concentrations ranging from 100 μg/mL up to 1400 μg/mL. Resistant clones selection could require from 1 to up to 3 weeks.
 | |
| Names | |
|---|---|
| Preferred IUPAC name
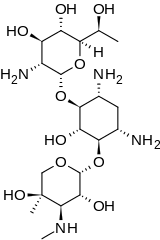
(2R,3S,4R,5R,6S)-5-Amino-6-{[(1R,2S,3S,4R,6S)-4,6-diamino-3-{[(2R,3R,4R,5R)-3,5-dihydroxy-5-methyl-4-(methylamino)oxan-2-yl]oxy}-2-hydroxycyclohexyl]oxy}-2-[(1R)-1-hydroxyethyl]oxane-3,4-diol | |
| Other names
Geneticin O-2-Amino-2,7-didesoxy-D-glycero-α-D-gluco-heptopyranosyl-(1→4)-O-(3-desoxy-4-C-methyl-3-(methylamino)-β-L-arabinopyranosyl- (1→6))-D-streptamin | |
| Identifiers | |
CAS Number |
|
3D model (JSmol) |
|
| ChEMBL | |
| ChemSpider | |
| DrugBank | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
InChI
| |
SMILES
| |
| Properties | |
Chemical formula |
C20H40N4O10 |
| Molar mass | 496.558 g·mol−1 |
Solubility in water |
50 mg/mL |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
G418 impurity profile
G418 is produced by fermentation and isolation processes and the G418 producing strain Micromonospora rhodorangea produces many other gentamicins while producing G418. The common impurities of G418 include gentamicins A, C1, C1a, C2, C2a and X2.[3] The quality of G418 is not based on just the potency, but more on the selectivity defined by the killing curve of the sensitive cells vs the resistant cells. A good G418 product has the lowest LD50 for sensitive cells (such as NIH 3T3) and the highest LD50 (can be up to 5,000 μg/ml) for resistant cells (NIH 3T3 transfected with resistant genes). Gentamicins have almost no selectivity, except gentamicin X2.[4]
Use in cell biology
G418 is routinely used as a selective agent in cell culture set-ups. Researchers can link the neoR selective resistance gene with their vector. Then if the vector is successfully introduced into cells, the cells can become G418-resistant cells. After treating with G418, these vector(-) cells will die, while vector(+) cells will survive. This method can help researchers select vector(+) cells.[1][5]
References
- "Geneticin". Thermo Fisher Scientific.
- "G418". labome.com. Archived from the original on 2009-12-29. Retrieved 2010-01-09.
- G418 impurity profile
- G418 selectivity
- Harvey Lodish; et al. (2013). "Chapter5: Molecular Genetic Techniques". Molecular Cell Biology (7th edition). Macmillan Higher Education. pp. 171–223. ISBN 978-1-4641-0981-2.