Oxidase test
The oxidase test is used to determine if an organism possesses the cytochrome c oxidase enzyme. The test is used as an aid for the differentiation of Neisseria, Moraxella, Campylobacter and Pasteurella species (oxidase positive). It is also used to differentiate pseudomonads from related species.[1]
Classification
Strains may be either oxidase-positive (OX+) or oxidase-negative (OX-).
OX+
OX+ normally means the bacterium contains cytochrome c oxidase (also known as Complex IV) and can therefore use oxygen for energy production by converting O2 to H2O2 or H2O with an electron transfer chain.
The Pseudomonadaceae are typically OX+.[1]
The Gram-negative diplococci Neisseria and Moraxella are oxidase-positive.[2]
Many Gram-negative, spiral curved rods are also oxidase-positive, which includes Helicobacter pylori, Vibrio cholerae, and Campylobacter jejuni.
Oxidase variable
Legionella pneumophila may be oxidase-positive.[3]
OX−
OX− normally means the bacterium does not contain cytochrome c oxidase and, therefore, either cannot use oxygen for energy production with an electron transfer chain or employs a different cytochrome for transferring electrons to oxygen.
Enterobacteriaceae are typically OX−.[4]
Mechanism

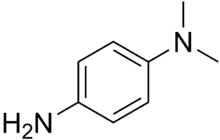
The test uses disks impregnated with a reagent such as N,N,N′,N′-tetramethyl-p-phenylenediamine, TMPD (or N,N-dimethyl-p-phenylenediamine, DMPD, which is also a redox indicator). The reagent is a dark-blue to maroon color when oxidized, and colorless when reduced. Oxidase-positive bacteria possess cytochrome oxidase or indophenol oxidase (an iron-containing hemoprotein).[5] These both catalyze the transport of electrons from donor compounds (NADH) to electron acceptors (usually oxygen). The test reagent TMPD acts as an artificial electron donor for the enzyme oxidase. The oxidized reagent forms the colored compound indophenol blue. The cytochrome system is usually only present in aerobic organisms that are capable of using oxygen as the terminal electron acceptor. The end-product of this metabolism is either water or hydrogen peroxide (broken down by catalase).[1]
Procedures
- Wet each disk with about four inoculating loops of deionized water.
- Use a loop to aseptically transfer a large mass of pure bacteria to the disk.
- Observe the disk for up to three minutes. If the area of inoculation turns dark-blue to maroon to almost black, then the result is positive. If a color change does not occur within three minutes, the result is negative.
In alternative manner, live bacteria cultivated on trypticase soy agar plates may be prepared using sterile technique with a single-line streak inoculation. The inoculated plates are incubated at 37 °C for 24–48 hours to establish colonies. Fresh bacterial preparations should be used. After colonies have grown on the medium, 2-3 drops of the reagent DMPD are added to the surface of each organism to be tested.
- A positive test (OX+) will result in a color change violet to purple, within 10–30 seconds.
- A negative test (OX-) will result in a light-pink or absence of coloration.
References
- MacFaddin JF, editor. Biochemical Tests for Identification of Medical Bacteria. 3rd ed. Philadelphia:Lippincott Williams and Wilkins; 2000. p. 363-7
- S. T., Cowan; Steel, K.J. (1993). Cowan and Steel's Manual for the Identification of Medical Bacteria (3rd ed.). Cambridge: Cambridge University Press. ISBN 9780511527104.
- "UK SMI" (PDF).
- Farmer JJ, Fanning GR, Huntley-Carter GP, et al. (May 1981). "Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp. nov. and Kluyvera cryocrescens sp. nov. in clinical specimens". J. Clin. Microbiol. 13 (5): 919–33. doi:10.1128/jcm.13.5.919-933.1981. PMC 273917. PMID 7240403.
- Isenberg HD, editor. Clinical Microbiology Procedures Handbook. American Society for Microbiology; 2004. p. 3.3.2-3.3.2.13.
- American Society for Microbiology, Oxidase Test Protocol. 2013. ASM MicrobeLibrary, 1–9.
- Cheng W J, Lin C W, Wu T G, Su C S, Hsieh M S. 2013. Calibration of glucose oxidase-based test strips for capillary blood measurement with oxygen saturated venous blood samples. Clinica Chimica Acta. 415, 152–157.
- Corchia L, Hubault R, Quinquenel B, N'Guyen. 2015. Rapidly Evolving Conjunctivitis Due to Pasteurella Multocida, Occurring after Direct Inoculation with Animal Droplets in an Immuno-compromised Host. BMC Ophthalmology 15.1, 21.
- Floch C, Alarcon-Gutiérrez E, Criquet S. 2007. ABTS assay of phenol oxidase activity in soil. Journal of Microbiological Methods. 71, 319–324.
- Gaby W L, Hadley C. 1957. Practical laboratory test for the identification of Pseudomonas aeruginosa. Journal of bacteriology. 74, 356–358.
- Gilani M, Munir T, Latif M, Gilani M, Rehman S, Ansari M, Hafeez A, Najeeb S, Saad N. 2015. In Vitro Efficacy of Doripenem against Pseudomonas Aeruginosa and Acinetobacter Baumannii by E-test. Journal of the College of Physicians and Surgeons Pakistan 25, 726–729.
- Kuss S, Tanner E E L, Ordovas-Montanes M, Compton R G. 2017. Electrochemical Recognition and Quantification of Cytochrome C Expression in and Aerobe/anaerobe Using, ','-tetramethyl—phenylene-diamine (TMPD). Chemical Science 8.11, 7682–7688. Web.
- Ivanova N V, Zemlak T S, Hanner R H, Hebert P D N. 2007. Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes. 7, 544–54.
- Prince C. 2009. Practical Manual of Medical Microbiology (Jaypee Brothers Medical Publishers (P) Ltd.) 112–112.
- Shields P, Cathcart L. 2013. Oxidase Test Protocol - Library. American Society for Microbiology, ASM MicrobeLibrary, 1–5.
- Steel K J. 1961. The Oxidase Reaction as a Taxonomic Tool. Journal of General Microbiology. 25, 297–306.
- Zanderigo F et al. 2018. [11C]Harmine Binding to Brain Monoamine Oxidase A: Test-Retest Properties and Noninvasive Quantification. Molecular Imaging and Biology. 20, 667–681.