Tetrahymena
Tetrahymena, a unicellular eukaryote, is a genus of free-living ciliates.[1] The genus Tetrahymena is the most widely studied member of its phylum.[2]: 59 It can produce, store and react with different types of hormones. Tetrahymena cells can recognize both related and hostile cells.[3]
| Tetrahymena | |
|---|---|
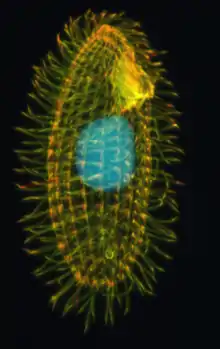 | |
| Tetrahymena thermophila | |
| Scientific classification | |
| Kingdom: | Chromista |
| Superphylum: | Alveolata |
| Phylum: | Ciliophora |
| Class: | Oligohymenophorea |
| Order: | Hymenostomatida |
| Family: | Tetrahymenidae |
| Genus: | Tetrahymena |
They can also switch from commensalistic to pathogenic modes of survival. They are common in freshwater lakes, ponds, and streams.[2]: 277
Tetrahymena species used as model organisms in biomedical research are T. thermophila and T. pyriformis.[4]
The most recent common ancestor to this genus was probably a small flagellated cell.[5]: 28
T. thermophila: a model organism in experimental biology
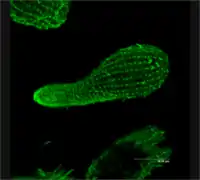
As a ciliated protozoan, Tetrahymena thermophila exhibits nuclear dimorphism: two types of cell nuclei. They have a bigger, non-germline macronucleus and a small, germline micronucleus in each cell at the same time and these two carry out different functions with distinct cytological and biological properties. This unique versatility allows scientists to use Tetrahymena to identify several key factors regarding gene expression and genome integrity. In addition, Tetrahymena possess hundreds of cilia and has complicated microtubule structures, making it an optimal model to illustrate the diversity and functions of microtubule arrays.
Because Tetrahymena can be grown in a large quantity in the laboratory with ease, it has been a great source for biochemical analysis for years, specifically for enzymatic activities and purification of sub-cellular components. In addition, with the advancement of genetic techniques it has become an excellent model to study the gene function in vivo. The recent sequencing of the macronucleus genome should ensure that Tetrahymena will be continuously used as a model system.
Tetrahymena thermophila exists in 7 different sexes (mating types) that can reproduce in 21 different combinations, and a single tetrahymena cannot reproduce sexually with itself. Each organism "decides" which sex it will become during mating, through a stochastic process.[6][7]
Studies on Tetrahymena have contributed to several scientific milestones including:
- First cell which showed synchronized division, which led to the first insights into the existence of mechanisms which control the cell cycle.[8]
- Identification and purification of the first cytoskeleton based motor protein such as dynein.[8]
- Aid in the discovery of lysosomes and peroxisomes.[8]
- Early molecular identification of somatic genome rearrangement.[8]
- Discovery of the molecular structure of telomeres, telomerase enzyme, the templating role of telomerase RNA and their roles in cellular senescence and chromosome healing (for which a Nobel Prize was won).[8]
- Nobel Prize–winning co-discovery (1989, in Chemistry) of catalytic RNA (ribozyme).[8][9]
- Discovery of the function of histone acetylation.[8]
- Demonstration of the roles of posttranslational modification such as acetylation and glycylation on tubulins and discovery of the enzymes responsible for some of these modifications (glutamylation)
- Crystal structure of 40S ribosome in complex with its initiation factor eIF1
- First demonstration that two of the "universal" stop codons, UAA and UAG, will code for the amino acid glutamine in some eukaryotes, leaving UGA as the only termination codon in these organisms.[10]

Life cycle
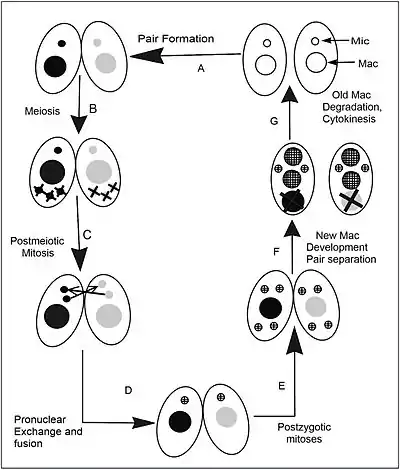
The life cycle of T. thermophila consists of an alternation between asexual and sexual stages. In nutrient rich media during vegetative growth cells reproduce asexually by binary fission. This type of cell division occurs by a sequence of morphogenetic events that results in the development of duplicate sets of cell structures, one for each daughter cell. Only during starvation conditions will cells commit to sexual conjugation, pairing and fusing with a cell of opposite mating type. Tetrahymena has seven mating types; each of which can mate with any of the other six without preference, but not its own.
Typical of ciliates, T. thermophila differentiates its genome into two functionally distinct types of nuclei, each specifically used during the two different stages of the life cycle. The diploid germline micronucleus is transcriptionally silent and only plays a role during sexual life stages. The germline nucleus contains 5 pairs of chromosomes which encode the heritable information passed down from one sexual generation to the next. During sexual conjugation, haploid micronuclear meiotic products from both parental cells fuse, leading to the creation of a new micro- and macronucleus in progeny cells. Sexual conjugation occurs when cells starved for at least 2hrs in a nutrient-depleted media encounter a cell of complementary mating type. After a brief period of co-stimulation (~1hr), starved cells begin to pair at their anterior ends to form a specialized region of membrane called the conjugation junction.

It is at this junctional zone that several hundred fusion pores form, allowing for the mutual exchange of protein, RNA and eventually a meiotic product of their micronucleus. This whole process takes about 12 hours at 30 °C, but even longer than this at cooler temperatures. The sequence of events during conjugation is outlined in the accompanying figure.[11]
The larger polyploid macronucleus is transcriptionally active, meaning its genes are actively expressed, and so it controls somatic cell functions during vegetative growth. The polyploid nature of the macronucleus refers to the fact that it contains approximately 200–300 autonomously replicating linear DNA mini-chromosomes. These minichromosomes have their own telomeres and are derived via site-specific fragmentation of the five original micronuclear chromosomes during sexual development. In T. thermophila each of these minichromosomes encodes multiple genes and exists at a copy number of approximately 45-50 within the macronucleus. The exception to this is the minichromosome encoding the rDNA, which is massively upregulated, existing at a copy number of approximately 10,000 within the macronucleus. Because the macronucleus divides amitotically during binary fission, these minichromosomes are un-equally divided between the clonal daughter cells. Through natural or artificial selection, this method of DNA partitioning in the somatic genome can lead to clonal cell lines with different macronuclear phenotypes fixed for a particular trait, in a process called phenotypic assortment. In this way, the polyploid genome can fine-tune its adaptation to environmental conditions through gain of beneficial mutations on any given mini-chromosome whose replication is then selected for, or conversely, loss of a minichromosome which accrues a negative mutation. However, the macronucleus is only propagated from one cell to the next during the asexual, vegetative stage of the life cycle, and so it is never directly inherited by sexual progeny. Only beneficial mutations that occur in the germline micronucleus of T. thermophila are passed down between generations, but these mutations would never be selected for environmentally in the parental cells because they are not expressed.[12]
Behavior
Free swimming cells of Tetrahymena are attracted to certain chemicals by chemokinesis. The major chemo-attractants are peptides and/or proteins.[13]
A 2016 study found that cultured Tetrahymena have the capacity to 'learn' the shape and size of their swimming space. Cells confined in a droplet of a water for a short time were, upon release, found to repeat the circular swimming trajectories 'learned' in the droplet. The diameter and duration of these swimming paths reflected the size of the droplet and time allowed to adapt.[14]
DNA repair
It is common among protists that the sexual cycle is inducible by stressful conditions such as starvation. Such conditions often cause DNA damage. A central feature of meiosis is homologous recombination between non-sister chromosomes. In T. thermophila this process of meiotic recombination may be beneficial for repairing DNA damages caused by starvation.
Exposure of T. thermophila to UV light resulted in a greater than 100-fold increase in Rad51 gene expression.[15] Treatment with the DNA alkylating agent methyl methanesulfonate also resulted in substantially elevated Rad 51 protein levels. These findings suggest that ciliates such as T. thermophila utilize a Rad51-dependent recombinational pathway to repair damaged DNA.
The Rad51 recombinase of T. thermophila is a homolog of the Escherichia coli RecA recombinase. In T. thermophila, Rad51 participates in homologous recombination during mitosis, meiosis and in the repair of double-strand breaks.[16] During conjugation, Rad51 is necessary for completion of meiosis. Meiosis in T. thermophila appears to employ a Mus81-dependent pathway that does not use a synaptonemal complex and is considered secondary in most other model eukaryotes.[17] This pathway includes the Mus81 resolvase and the Sgs1 helicase. The Sgs1 helicase appears to promote the non-crossover outcome of meiotic recombinational repair of DNA,[18] a pathway that generates little genetic variation.
Phenotypic and Genotypic Plasticity
Many species of Tetrahymena are known to display unique response mechanisms to stress and various environmental pressures. The unique genomic architecture of the ciliates (presence of a MIC, high ploidy, large number of chromosomes, etc.) allows for differential gene expression, as well as increased genomic flexibility. The following is a non-exhaustive list of examples of phenotypic and genotypic plasticity in the Tetrahymena genus.
Inducible Trophic Polymorphisms
T. vorax is known for its inducible trophic polymorphisms, an ecologically offensive tactic that allows it to change its feeding strategy and diet by altering its morphology.[19] Normally, T. vorax is a bacterivorous microstome around 60 μm in length. However, it has the ability to switch into a carnivorous macrostome around 200 μm in length that can feed on larger competitors. If T. vorax cells are too nutrient starved to undertake transformation, they have also been recorded as transforming into a third “tailed”-microstome morph, thought to be a defense mechanism in response to cannibalistic pressure. While T. vorax is the most well studied Tetrahymena that exhibits inducible trophic polymorphisms, many lesser known species are able to undertake transformation as well, including T. paulina and T. paravorax.[20] However, only T. vorax has been recorded as having both a macrostome and tailed-microstome form.
These morphological switches are triggered by an abundance of stomatin in the environment, a mixture of metabolic compounds released by competitor species, such as Paramecium, Colpidium, and other Tetrahymena. Specifically, chromatographic analysis has revealed that ferrous iron, hypoxanthine, and uracil are the chemicals in stomatin responsible for triggering the morphological change.[21] Many researchers cite “starvation conditions” as inducing the transformation, as in nature, the compound inducers are in highest concentration after microstomal ciliates have grazed down bacterial populations, and ciliates populations are high. When the chemical inducers are in high concentration, T. vorax cells will transform at higher rates, allowing them to prey on their former trophic competitors.
The exact genetic, and structural mechanisms that underlie T. vorax transformation are unknown. However, some progress has been made in identifying candidate genes. Researchers from the University of Alabama have used cDNA subtraction to remove actively transcribed DNA from microstome and macrostome T. vorax cells, leaving only differentially transcribed cDNA molecules.[22] While nine differentiation-specific genes were found, the most frequently expressed candidate gene was identified as a novel sequence, SUBII-TG.
The sequenced region of SUBII-TG was 912 bp long and consists of three largely identical 105 bp open-reading frames. A northern blot analysis revealed that low levels of transcription are detected in microstome cells, while high levels of transcription occur in macrostome cells. Furthermore, when the researchers limited SUBII-TG expression in the presence of stomatin (using antisense oligonucleotide methods), a 55% reduction in SUBII-TG mRNA correlated with a 51% decrease in transformation, supporting the notion that the gene is at least partially responsible for controlling the transformation in T. vorax. However, very little is known about the SUBII-TG gene. Researchers were only able to sequence a portion of the entire open-reading frame, and other candidate genes have not been investigated thoroughly. mRNA and amino acid sequencing indicate that ubiquitin may play a crucial role in allowing transformation to take place as well. However, no known genes in the ubiquitin family have been identified in T. vorax.[23] Finally, the genetic mechanisms of the “tailed” microstome morph are completely unknown.
Metal Resistance, Gene & Genome Amplification
Other related species exhibit their own unique responses to various stressors. In T. thermophila, chromosome amplification and gene expansion are inducible responses to common organometallic pollutants such as Cadmium, Copper, and Lead.[24] Strains of T. thermophila that were exposed to large quantities of Cd2+ over time were found to have a 5-fold increase of MTT1, and MTT3 (metallothionein genes that code for Cadmium and Lead binding proteins) as well as CNBDP, an unrelated gene that lies just upstream of MTT1 on the same chromosome. The fact that a non-metallothionein gene on the same locus as MTT1 and MTT3 increased copy number indicates that the entire chromosome had been amplified, as opposed to just specific genes. Tetrahymena species are 45-ploid for their macronucleus, meaning that the wild type of T. thermophila normally contains 45 copies of each chromosome. While the actual number of unique chromosomes are unknown, the number is thought to be around 187 in the MAC, and 5 in the MIC.[25] Thus, the Ca2+ adapted strain contained 225 copies of the specific chromosome in question. This resulted in a nearly 28 times increase in detected expression levels of MTT1, and slightly less in MTT3.
Interestingly, when researchers grew a sample of the T. thermophila population in normal growth medium (lacking Cd2+) for 1 month, the number of MTT1, MTT3, and CNBDP genes decreased to an average of 3 copies (135C). By 7 months in normal growth medium, the T. thermophila cells were found reduced to just the wild type copy number (45C). When researchers returned cells from the same colony to Cd2+ medium, within a week MTT1, MTT3, and CNBDP genes increased to 3 copies once again (135C). Thus, the authors argue that chromosome amplification is an inducible and reversible mechanism in the Tetrahymena genetic response to metal stress.
Researchers also used gene-knockdown experiments, where the copy number of another metallothionein gene on a different chromosome, MTT5, was dramatically reduced. Within a week, the new strain was found to have developed 4 novel genes from at least 1 duplication of MTT1. However, chromosome duplication had not taken place, as indicated by the wild-type ploidy and the normal quantity of other genes on the same chromosomes. Rather, researchers believe that the duplication resulted from homologous recombination events, producing transcriptionally active, upregulated genes that carry repeated MTT1.
Enhanced Motility and Dispersal
T. thermophila also undergoes phenotypic changes when faced with limited resource availability. Cells are capable of changing their shape and size, along with behavioral swimming strategies in response to starvation.[26] The more motile cells that change in response to starvation are known as dispersers, or disperser cells. While rates and levels of phenotypic change differ between strains, disperser cells form in nearly all strains of T. thermophila when faced with starvation. Dispersers, and non-dispersing cells both become dramatically thinner and smaller, increasing the basal body and cilia density, allowing them to swim between 2 to 3 times faster than normal cells.[27] Some strains of T. thermophila have also been found to develop a single, non-beating, enlarged cilia that assists the cell in steering or directing movement. While the behavior has been shown to correlate with faster dispersal and form as a reversible trait in Tetrahymena cells, little is known about the genetic or cellular mechanisms that allow for its development. Furthermore, other studies show that when genetically variable populations of T. thermophila were starved, dispersal cells actually increased in cell length, despite still becoming thinner.[28] More research is needed to determine the genetic mechanisms that underlie disperser formation.
Species in genus
Species in this genus include.[1]
- Tetrahymena americanis
- Tetrahymena asiatica
- Tetrahymena australis
- Tetrahymena bergeri
- Tetrahymena borealis
- Tetrahymena canadensis
- Tetrahymena capricornis
- Tetrahymena caudata
- Tetrahymena chironomi
- Tetrahymena corlissi
- Tetrahymena cosmopolitanis
- Tetrahymena dimorpha
- Tetrahymena edaphoni
- Tetrahymena elliotti
- Tetrahymena empidokyrea
- Tetrahymena farahensis
- Tetrahymena farleyi
- Tetrahymena furgasoni
- Tetrahymena glochidiophila
- Tetrahymena hegewischi
- Tetrahymena hyperangularis
- Tetrahymena leucophrys
- Tetrahymena limacis
- Tetrahymena lwoffi
- Tetrahymena malaccensis
- Tetrahymena mimbres
- Tetrahymena mobilis
- Tetrahymena nanneyi
- Tetrahymena nipissingi
- Tetrahymena paravorax
- Tetrahymena patula
- Tetrahymena pigmentosa
- Tetrahymena pyriformis
- Tetrahymena rostrata
- Tetrahymena rotunda
- Tetrahymena setifera
- Tetrahymena setigera
- Tetrahymena setosa
- Tetrahymena shanghaiensis
- Tetrahymena sialidos
- Tetrahymena silvana
- Tetrahymena skappus
- Tetrahymena sonneborni
- Tetrahymena stegomyiae
- Tetrahymena thermophila
- Tetrahymena tropicalis
- Tetrahymena vorax
References
- "Tetrahymena - Encyclopedia of Life". eol.org. Retrieved 2021-10-16.
- Tetrahymena Thermophila. Academic Press. 2012-10-22. ISBN 978-0-12-385968-6.
- Csaba, György (September 2016). "Lectins and Tetrahymena – A review". Acta Microbiologica et Immunologica Hungarica. 63 (3): 279–291. doi:10.1556/030.63.2016.001. ISSN 1217-8950. PMID 27539329.
- Elliott AM (1973). Biology of Tetrahymena. Dowen, Hutchinson and Ross Inc. ISBN 978-0-87933-013-2.
- Tetrahymena Thermophila. Academic Press. 1999-09-21. ISBN 978-0-08-057362-5.
- Cervantes MD, Hamilton EP, Xiong J, Lawson MJ, Yuan D, Hadjithomas M, et al. (2013). "Selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila". PLOS Biology. 11 (3): e1001518. doi:10.1371/journal.pbio.1001518. PMC 3608545. PMID 23555191.
- Quirk, Trevor (2013-03-27). "How a microbe chooses among seven sexes". Nature: nature.2013.12684. doi:10.1038/nature.2013.12684. ISSN 0028-0836. S2CID 179307103.
- Orias E (10 February 2002). "Sequencing the Tetrahymena thermophila Genome White Paper". National Human Genome Research Institute.
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (November 1982). "Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena". Cell. 31 (1): 147–57. doi:10.1016/0092-8674(82)90414-7. PMID 6297745. S2CID 14787080.
- Horowitz S, Gorovsky MA (April 1985). "An unusual genetic code in nuclear genes of Tetrahymena". Proceedings of the National Academy of Sciences of the United States of America. 82 (8): 2452–5. Bibcode:1985PNAS...82.2452H. doi:10.1073/pnas.82.8.2452. PMC 397576. PMID 3921962.
- Elliott, AM; Hayes, RE (1953). "Mating Types in Tetrahymena". Biological Bulletin. 105 (2): 269–284. doi:10.2307/1538642. JSTOR 1538642.
- Prescott DM (June 1994). "The DNA of ciliated protozoa". Microbiological Reviews. 58 (2): 233–67. doi:10.1128/MMBR.58.2.233-267.1994. PMC 372963. PMID 8078435.
- Leick V, Hellung-Larsen P (January 1992). "Chemosensory behaviour of Tetrahymena". BioEssays. 14 (1): 61–6. doi:10.1002/bies.950140113. PMID 1546982.
- Kunita I, Yamaguchi T, Tero A, Akiyama M, Kuroda S, Nakagaki T (May 2016). "A ciliate memorizes the geometry of a swimming arena". Journal of the Royal Society, Interface. 13 (118): 20160155. doi:10.1098/rsif.2016.0155. PMC 4892268. PMID 27226383.
- Campbell C, Romero DP (July 1998). "Identification and characterization of the RAD51 gene from the ciliate Tetrahymena thermophila". Nucleic Acids Research. 26 (13): 3165–72. doi:10.1093/nar/26.13.3165. PMC 147671. PMID 9628914.
- Marsh TC, Cole ES, Stuart KR, Campbell C, Romero DP (April 2000). "RAD51 is required for propagation of the germinal nucleus in Tetrahymena thermophila". Genetics. 154 (4): 1587–96. doi:10.1093/genetics/154.4.1587. PMC 1461009. PMID 10747055.
- Chi J, Mahé F, Loidl J, Logsdon J, Dunthorn M (March 2014). "Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway". Molecular Biology and Evolution. 31 (3): 660–72. doi:10.1093/molbev/mst258. PMID 24336924.
- Lukaszewicz A, Howard-Till RA, Loidl J (November 2013). "Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex". Nucleic Acids Research. 41 (20): 9296–309. doi:10.1093/nar/gkt703. PMC 3814389. PMID 23935123.
- Banerji, Aabir; Morin, Peter J. (May 2014). "Trait-mediated apparent competition in an intraguild predator-prey system". Oikos. 123 (5): 567–574. doi:10.1111/j.1600-0706.2013.00937.x.
- Ryals, Phillip E.; Smith-Somerville, Harriett E.; Buhse, Howard E. (2002-01-01), Jeon, Kwang W. (ed.), "Phenotype Switching in Polymorphic Tetrahymena: A Single-Cell Jekyll and Hyde", International Review of Cytology, A Survey of Cell Biology, Academic Press, 212: 209–238, doi:10.1016/s0074-7696(01)12006-1, ISBN 9780123646163, PMID 11804037, retrieved 2022-05-11
- Smith-Somerville, Harriett E.; Hardman, John K.; Timkovich, Russell; Ray, William J.; Rose, Karen E.; Ryals, Phillip E.; Gibbons, Sandra H.; Buhse, Howard E. (2000-06-20). "A complex of iron and nucleic acid catabolites is a signal that triggers differentiation in a freshwater protozoan". Proceedings of the National Academy of Sciences. 97 (13): 7325–7330. Bibcode:2000PNAS...97.7325S. doi:10.1073/pnas.97.13.7325. ISSN 0027-8424. PMC 16544. PMID 10860998.
- Green, M. M.; LeBoeuf, R. D.; Churchill, P. F. (2000). "Biological and molecular characterization of cellular differentiation in Tetrahymena vorax: a potential biocontrol protozoan". Journal of Basic Microbiology. 40 (5–6): 351–361. doi:10.1002/1521-4028(200012)40:5/6<351::aid-jobm351>3.0.co;2-q. ISSN 0233-111X. PMID 11199495. S2CID 21981461.
- "Analysis of ubiquitin and differential gene expression during differentiation in Tetrahymena vorax - ProQuest". www.proquest.com. ProQuest 304234889. Retrieved 2022-05-11.
- de Francisco, Patricia; Martín‐González, Ana; Turkewitz, Aaron P.; Gutiérrez, Juan Carlos (July 2018). "Genome plasticity in response to stress in Tetrahymena thermophila : selective and reversible chromosome amplification and paralogous expansion of metallothionein genes". Environmental Microbiology. 20 (7): 2410–2421. doi:10.1111/1462-2920.14251. ISSN 1462-2912. PMC 6117198. PMID 29687579.
- Yao, Meng-Chao; Chao, Ju-Lan; Cheng, Chao-Yin (2014-11-21). Gellert, Martin; Craig, Nancy (eds.). "Programmed Genome Rearrangements in Tetrahymena". Microbiology Spectrum. 2 (6): 2.6.31. doi:10.1128/microbiolspec.MDNA3-0012-2014. ISSN 2165-0497. PMID 26104448.
- Junker, Anthony D.; Jacob, Staffan; Philippe, Hervé; Legrand, Delphine; Pearson, Chad G. (2021-08-20). "Plastic cell morphology changes during dispersal". iScience. 24 (8): 102915. Bibcode:2021iSci...24j2915J. doi:10.1016/j.isci.2021.102915. ISSN 2589-0042. PMC 8367785. PMID 34430806.
- Fjerdingstad, Else J.; Schtickzelle, Nicolas; Manhes, Pauline; Gutierrez, Arnaud; Clobert, Jean (2007-08-06). "Evolution of dispersal and life history strategies – Tetrahymena ciliates". BMC Evolutionary Biology. 7 (1): 133. doi:10.1186/1471-2148-7-133. ISSN 1471-2148. PMC 1997130. PMID 17683620.
- Jacob, Staffan; Laurent, Estelle; Morel‐Journel, Thibaut; Schtickzelle, Nicolas (February 2020). "Fragmentation and the context‐dependence of dispersal syndromes: matrix harshness modifies resident‐disperser phenotypic differences in microcosms". Oikos. 129 (2): 158–169. doi:10.1111/oik.06857. ISSN 0030-1299. S2CID 208584362.
Further reading
- Asai DJ, Forney JD, eds. (2000). Tetrahymena thermophila. Methods in Cell Biology. Vol. 62. Academic Press. ISBN 978-0-12-544164-3.
- Collins K, Gorovsky MA (May 2005). "Tetrahymena thermophila". Current Biology. 15 (9): R317-8. doi:10.1016/j.cub.2005.04.039. PMID 15886083. S2CID 11738486.
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, et al. (September 2006). "Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote". PLOS Biology. 4 (9): e286. doi:10.1371/journal.pbio.0040286. PMC 1557398. PMID 16933976.
External links
- Tetrahymena Stock Center at Cornell University
- ASSET: Advancing Secondary Science Education thru Tetrahymena
- Tetrahymena Genome Database
- Biogeography and Biodiversity of Tetrahymena
- Tetrahymena thermophila Genome Project at The Institute for Genomic Research
- Tetrahymena thermophila Genome Sequence Synopsis
- Tetrahymena thermophila genome paper
- Tetrahymena experiments on Journal of Visualized Experiments (JoVE) website
- Microbial Digital Specimen Archives: Tetrahymena image gallery
- All Creatures Great and Small: Elizabeth Blackburn