Zona limitans intrathalamica
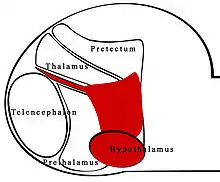
The zona limitans intrathalamica (ZLI) is a lineage-restriction compartment and primary developmental boundary in the vertebrate forebrain (which is analogous to the human cerebrum) that serves as a signaling center and a restrictive border between the thalamus (also known as the dorsal thalamus) and the prethalamus (ventral thalamus).
Sonic hedgehog (shh) signaling from the ZLI is crucial in the development of the diencephalon,[1] which develops into the thalamus, the pretectum, and the anterior tegmental structures.[2] The ZLI together with the prethalamus and thalamus make up the mid-diencephalic territory (MDT).[3]
Discovery
Cell lineage restriction boundaries, across which replicating cells cannot migrate, were first discovered in invertebrates, where the expression of various Hox genes in each compartment confer the differentiation of observable segments in the adult body of Drosophila melanogaster. Analogous structures were discovered in the developing vertebrate brain. Rhombomeres, which extend down the embryo from the hindbrain, contain clear boundaries and each express various Hox genes that are necessary for differentiation of structures within the body. More anterior regions of the brain were examined in search of other cell lineage restriction boundaries, and multiple potential boundaries continue to be studied (see Developmental Boundaries).[4]
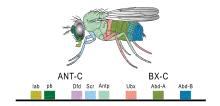
The importance of these compartments as local signaling centers, areas which chemically influence surrounding tissue, was elucidated by first observing differential expression of Hox genes in various compartments and second by observing mutant D. melanogaster and corresponding phenotypic (physical) changes.[4]
The ZLI was first discovered in the chick using explant and lineage-labeling experiments. In the explant experiments, cells in the region that will become the ZLI, the prethalamus, and the thalamus in the chick were removed and placed into separate cultures; the cells continued to grow and retained their identity (the ZLI began transcribing shh, whereas the prethalamus and thalamus did not). Necessity of the ZLI and its corresponding shh expression were evidenced by lack of thalamic and prethalamic genetic markers in culture (see Signaling).[5] These experiments confirmed the ZLI as a signaling center. In lineage-labeling experiments, cells were genetically marked, so that each time a labeled cell replicated, its progeny were marked as well. Cells that were marked in the developing ZLI and their progeny remained restricted to the zone. These experiments demonstrated the ZLI as a cell-lineage restriction boundary.[6]
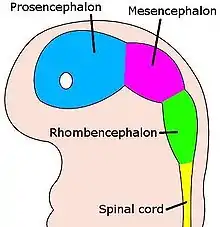
Not only a boundary, the ZLI is also a compartment with separate cell lineage restriction boundaries both anterior and posterior of a section of shh expression. The importance of the ZLI was once more confirmed by ectopic expression of shh in other regions of the forebrain, known as the prosencephalon during development (both the telencephalon and diencephalon), inducing a ZLI-like region that induces thalamic fate.[7]
Developmental boundaries
During development in both vertebrates and invertebrates, cell lineage restriction boundaries and signaling centers are formed in order to ensure proper differentiation of the body. Chemical signals, like shh from the ZLI, are often released from these boundaries and compartments in a concentration gradient (the chemicals are in much higher concentrations closer to the source) and confer identity to flanking regions. Other genes differentially expressed in these flanking regions aid in ensuring proper differentiation (see Signaling).
Many developmental boundaries have been studied: within the forebrain alone, the confirmed cell lineage restriction boundaries are the pallial-subpallial boundary (PSB) dividing the dorsal and ventral telencephalon, the diencephalon-midbrain boundary (DMB) posterior to the ZLI, and the ZLI. The ZLI, like each rhombomere, serves as an independent compartment that confers the identity of diencephalon in anterior and posterior regions. Other developmental boundaries serve as cell-lineage restriction boundaries but not signaling centers, while others are signaling centers to and from which cells can migrate. Despite discoveries of cell lineage restriction boundaries and compartments in the brain, many of the regions studied have been disproven as segmental boundaries. These areas have potential as signaling centers, which have influence over the development of neighboring tissues.[4]
These boundaries have great influence over other regions of the brain: the placement of the ZLI not only affects the size of adjacent regions but also the size of the telencephalon. A posterior shift in the ZLI allows more cells to be allocated to the telencephalon. The same is true for other developmental boundaries in the brain and throughout the body: shifts in boundaries responsible for allocating a certain amount of tissue to a certain function result in drastic changes in the adult structure. These boundaries are of crucial importance for proper differentiation.
Formation
Initial axis patterning
After gastrulation, the embryo is completely undifferentiated and requires many different cues to initiate proper differentiation of the body. The top (roof plate, on the dorsal side of the embryo) and bottom (floor plate, on the ventral side) play a crucial role in these first steps: each acts through global signaling (signaling throughout the entire embryo) for dorsoventral neural patterning. After completion of the development of the dorsoventral axis, more local signaling occurs in the developing brain: developmental boundaries such as the midbrain-hindbrain boundary (MHB), rhombomeres, and the ZLI aid in anteroposterior organization.[5]
Emergence of Shh expression
Shortly after the beginning of dorsoventral patterning, Shh is expressed along the basal plate (the bottom) of the embryo, which functions in "ventralization of the neural tube, promotion of growth and proliferation, and formation of the hypothalamus".[8] As the embryo continues to develop, Shh expression characteristic of the ZLI extends dorsally to form a wedge that eventually narrows to a strip at approximately 22 somites (the number of developed rhombomeres) or less than one day in zebrafish. Although Shh expression extends dorsally from the basal plate, the ZLI is capable of forming even without the basal plate or mesodermal tissue. Shh cooperates with dlx2 and fezl anteriorly and IRX3 and dbx1a posteriorly (genes expressed vary among different organisms), which are genes that are expressed in the prethalamus and thalamus, respectively.[3]
The ZLI is also characterized by a lack of lunatic fringe (lfng), which is observable even before Shh expression is observed in the forming ZLI. This indicates that cells are fated for ZLI formation before induction by Shh (n.b. Shh]' signals from the ZLI and is characteristic of the compartment during and after formation). Lfng is expressed very early (soon after gastrulation) in the region that will become the ZLI, but shortly thereafter expression recedes through migration of lfng-expressing cells to form the lfng-free wedge characteristic of the developing ZLI. Shh expression does not extend dorsally until a few hours later.[5]
Positioning
The factors influencing the formation and location of the ZLI are widely studied but still disputed. Differences between different animal models further complicate elucidation of the genetic pathways, specifically between chicks/ mammalian models and zebrafish.
Wnt (the family wingless) genes are crucial for the development of the ZLI both directly and indirectly in all animal systems. Along with the role of Wnt genes in patterning the anteroposterior axis through gradient polarization, Wnt8b is expressed within the ZLI itself and may help guide dorsal movement of Shh expression.[9] The Wnt polarization gradient has been linked to induction of ZLI-patterning genes IRX3 and SIX3, which border the ZLI posteriorly and anteriorly, respectively. However, these genes have been shown to be non-essential for ZLI formation in zebrafish and have been reevaluated in other models.[1][3]
Specification of the ZLI may also involve the ventral tissues of the embryo: the prechordal and epichordal plates, or neuroepithelial tissue (see figure). Interactions between these tissues may be responsible for the recession of expression of lfng which allows dorsal movement Shh expression.[10] The specific prechordal and epichordal plates, characterized by expression of SIX3 and IRX3, respectively, may influence positioning of the ZLI more so than the genes themselves.[5] The prechordal plate ventrally borders the telencephalon, with the epichordal plate posterior to it.
Studies of the formation of the ZLI performed in zebrafish have revealed the significance of OTX2 and IRX1 in ZLI positioning. OTX2 expression characterizes the developing optic tectum, which is responsible for sight processing. Expression extends anteriorly and ends sharply at the ZLI, with high expression along the line where Shh is expressed.[11] Before the ZLI is formed, OTX2] is expressed ubiquitously throughout the forebrain, and begins to recede to the position of the putative ZLI. Experiments where OTX2] expression was repressed showed no dorsal movement of Shh expression and no ZLI formation.[3] Irx1, which is analogous to IRX3 in birds and mammals, is expressed posterior to the ZLI. Studies have suggested that while OTX2 positively restricts the ZLI anteriorly (Shh cannot be expressed where OTX2 is not), 'IRX1 negatively restricts the ZLI posteriorly.[3] Other genes crucial for differentiation in the brain, including Fgf genes responsible for patterning of the midbrain-hindbrain boundary (MHB), have been implicated in positioning of the ZLI.[9]
Studies on the role of Shh signaling in the ZLI were difficult to study for many years, because mutants lacking expression have many developmental deficits including lack of a diencephalon.[12] Explant and lineage-labeling experiments previously described aided in elucidation of the role of Shh and other genes in differentiation of these tissues. More recently, the mouse Shh;Gli3 double mutant was found to have an enlarged diencephalon with a ring of Fgf8 and Wnt in place of the ZLI, indicating a complex interaction between Shh and these genes at the ZLI.[13] This also indicates that other patterning cues are able to establish Fgf8 and Wnt signaling domains at the ZLI in the absence of Shh and Gli3.
Differentiation after ZLI degradation
After differentiation of the progenitor cells (at a precise stage yet to be fully determined), the ZLI and its lineage restriction disappears, allowing cells to migrate across the former boundary and the dorsal and ventral thalami to merge into one functional unit, as shown by replication-incompetent retroviral experiments that marked cells and showed their migration throughout the diencephalon.[9]
Signaling
After establishment of the ZLI, shh has been shown to induce expression of thalamic and prethalamic markers, gbx2 and dlx2/ nkx2.1, respectively. This differential induction most likely is due to the expression of genes such as IRX3 in the thalamus: ectopic expression experiments showed that if IRX3, which is normally expressed in the developing thalamus, is expressed anterior to the ZLI, then the developing prethalamus will change identity.[10] Note that these genes which help confer shh competence help to pattern the ZLI.
Signaling from the ZLI cooperating with thalamic and prethalamic markers ensures the migration of post-mitotic (neural progenitor) cells to the mantle zone where the cells assemble into nuclei characteristic of the thalamus. These nuclei are the mechanism of the relay of information from the thalamus to the cortex. The thalamus itself is highly diverse, with each nucleus having distinct morphologies and physiologies according to the region of the brain to which it is connected. These differences are thought to originate in differential gene expression in the thalamus and prethalamus, which allows for a single structure with multiple different and separate functions once the two have merged and completed growth and differentiation.[9] So a functional interplay between Shh from the ZLI and the bHLH factor Her6 (homolog to HES1) determines the neuronal identity within the thalamus: Her6 positive cells in the prethalamus and rostral thalamus differentiate into GABAergic inhibitory neurons, whereas her negative cells become glutamatergic relay neurons. Both cell types depend on Shh signal as trigger to initiate the developmental programme.[14]
References
- Scholpp S, Lumsden A (Aug 2010). "Building a bridal chamber: development of the thalamus". Trends Neurosci. 33 (8): 373–380. doi:10.1016/j.tins.2010.05.003. PMC 2954313. PMID 20541814.
- Garcia-Lopez R, Vieira C, Echevarria D, Martinez S (2004). "Fate map of the diencephalon and the zona limitans at 10-somites stage in chick embryos". Developmental Biology. 268 (2): 514–530. doi:10.1016/j.ydbio.2003.12.038. PMID 15063186.
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C (2007). "Otx1l, Otx2, and Irx1b establish and position the ZLI in the diencephalon". Development. 134 (17): 3167–3176. doi:10.1242/dev.001461. PMC 7116068. PMID 17670791.
- Kiecker C, Lumsden A (2005). "Compartments and their boundaries in vertebrate brain development". Nature Reviews Neuroscience. 6 (7): 553–564. doi:10.1038/nrn1702. PMID 15959467.
- Guinazu MF, Chambers D, Lumsden A, Kiecker C (2007). "Tissue interactions in the developing chick diencephalon". Neural Development. 2 (25): 1–15. doi:10.1186/1749-8104-2-25. PMC 2217525. PMID 17999760.
- Zeltser LM, Larsen CW, Lumsden A (2001). "A new developmental compartment in the forebrain regulated by Lunatic fringe". Nature Neuroscience. 4 (7): 683–684. doi:10.1038/89455. PMID 11426219.
- Puelles L, Rubenstein JL (1993). "Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggests a neuromeric organization". Trends in Neurosciences. 16 (11): 472–479. doi:10.1016/0166-2236(93)90080-6. PMID 7507621.
- Scholpp S, Wolf O, Brand M, Lumsden A (2005). "Hedgehog signaling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon". Development. 133 (5): 855–864. doi:10.1242/dev.02248. PMID 16452095.
- Lim Y, Golden JA (2007). "Patterning the developing diencephalon". Brain Research Reviews. 53 (1): 17–26. doi:10.1016/j.brainresrev.2006.06.004. PMID 16876871.
- Vieira C, Garda AL, Shimamura K, Martinez S (2005). "Thalamic development induced by Shh in the chick embryo". Developmental Biology. 284 (2): 351–363. doi:10.1016/j.ydbio.2005.05.031. PMID 16026780.
- Ba-Charvet KT, von Boxberg Y, Guazzi S, Boncinelli E, Godement P (1998). "A potential role for the OTX2 homeoprotein in creating early 'highways' for axon extension in the rostral brain". Development. 125 (21): 4273–4282. PMID 9753681.
- Hashimoto-Torii K, Motoyama J, Hui CC, Kuroiwa A, Nakafuku M, Shimamura K (2003). "Differential activities of Sonic hedgehog mediated by Gli transcription factors define distinct neuronal subtypes in the dorsal thalamus". Mechanisms of Development. 120 (10): 1097–1111. doi:10.1016/j.mod.2003.09.001. PMID 14568100.
- Rash, BG; Grove, EA (15 November 2011). "Shh and Gli3 regulate formation of the telencephalic-diencephalic junction and suppress an isthmus-like signaling source in the forebrain". Developmental Biology. 359 (2): 242–50. doi:10.1016/j.ydbio.2011.08.026. PMC 3213684. PMID 21925158.
- Scholpp S, Delogu A, Gilthorpe J, Peukert D, Schindler S, Lumsden A. Her6 regulates the neurogenetic gradient and neuronal identity in the thalamus. Proc Natl Acad Sci U S A. 2009 Nov 24;106(47):19895-900