Enteroaggregative Escherichia coli
| Enteroaggregative Escherichia coli | |
|---|---|
 | |
| EAEC infecting human cells | |
| Specialty | Infectious disease |
| Symptoms | Diarrhea, mucoid diarrhea, fever, nausea, tenesmus[1] |
| Causes | pathotype of Escherichia coli [1][2] (EAEC is transmitted through the fecal-oral route and primarily contaminated by food and water.[3]) |
| Diagnostic method | Presence of EAEC-associated genes using PCR[4] |
| Treatment | Antibiotics (Cipro)[5][6] |
Enteroaggregative Escherichia coli (EAEC or EAggEC) are a pathotype of Escherichia coli which cause acute and chronic diarrhea in both the developed and developing world.[2][1] They may also cause urinary tract infections.[1] EAEC are defined by their "stacked-brick" pattern of adhesion to the human laryngeal epithelial cell line HEp-2.[7] The pathogenesis of EAEC involves the aggregation of and adherence of the bacteria to the intestinal mucosa, where they elaborate enterotoxins and cytotoxins that damage host cells and induce inflammation that results in diarrhea.
EAEC is now recognized as an emerging enteric pathogen. In particular, EAEC are reported as the second most common cause of traveler's diarrhea, second only to Enterotoxigenic E. coli, and a common cause of diarrhea amongst pediatric populations.[8][9] It has also been associated with chronic infections in the latter, as well as in immunocompromised hosts, such as HIV-infected individuals.[10] Awareness of EAEC was increased by a serious outbreak in Germany during 2011, causing about 3000 cases and at least 50 fatalities. The pathogen responsible was found to be an EAEC O104:H4 strain which was lysogenized by a Shiga toxin encoding phage (typically associated with Shiga toxin-producing Escherichia coli, which often encode the adhesin intimin).[11][12][13] The putative cause of the outbreak were sprouted fenugreek seeds.[14]
Strains of EAEC are highly genetically heterogeneous, and the identification of virulence factors important for pathogenesis has proven difficult.[4] Many EAEC encode a transcriptional factor named aggR (aggregative regulator), part of the AraC family of transcription activators. AggR regulates many plasmid, as well chromosomally encoded, virulence factors, that include genes implicated in aggregative adherence fimbriae biogenesis and toxin production. Several toxins have been linked to EAEC virulence, including ShET1 (Shigella enterotoxin 1), Pet (plasmid‐encoded toxin), and EAST-1. However, further studies of these factors have failed to elucidate their role in pathogenesis.[15]
Classification
Diarrhea is still an important disease burden worldwide. It causes considerable childhood mortality in the developing world and is correlated with morbidity (or of relating to disease) and substation health care costs in industrialized countries. The cause of infectious diarrhea is diarrheagenic Escheriachia coli (DEC) group. Subgroups of diarrheagenic Escheriachia coli (DEC) are the following: enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), enterotoxigenic E.coli (ETEC), Shiga toxin-producing E. coli (STEC) and Enteroaggregative E. coli (EAEC).[1]
E. coli is a bacterium that is found in the intestines, its mostly harmless, but some strains of bacteria can cause illness and infection,[16]
Signs and symptoms
Enteroaggregative Escheichia coli (EAEC) is a type of strain from E.coli. E.coli causes intestinal infections, some intestinal infections include diarrhea, fever and abdominal pain. Most severe cases can lead to bloody diarrhea, dehydration or even kidney failure. People with weakened immune systems, young children, older adults and pregnant women are at increased risks for developing these complications. Symptoms of intestinal infection usually begin between 8 and 52 hours after you have been infected with E.coli,[1] this is the incubation period.
The incubation period is the time between catching an infection and symptoms appearing.[17]
Bloody diarrhea has only been observed in children, and only rarely.[1] On the other hand, the STEC-EAEC hybrid strain identified in the 2011 Germany outbreak caused bloody diarrhea.[18]
Transmission
The trasmission of Enteroaggregative Escherichia coli can be:
- Contaminated water – Human and animal feces may pollute ground and surface water, and water used or irrigate crops. Although public water systems use chlorine and other chemicals to kill such organisms like E. coli, some outbreaks have been linked to contaminated water supplies.[19]
- Contaminated food – most common way to get a E.coli infection is by eating contaminated food such as ground beef, unpasteurized milk and fresh produce.[19]
- Improper food handling – by consuming raw food, or not cooking the food properly, especially meats and poultry. It can also be transmitted by not cleaning your cooking utensils properly, causing cross contamination.[20]
Pathogenesis
.jpg.webp)
EAEC is transmitted through the fecal-oral route and primarily contaminated by food and water.[3] EAEC has been associated with many symptoms such as diarrhea in some individuals and intestinal colonization in others.[5] Because many strains of EAEC have been identified, it is difficult to identify the mechanism of its pathogenesis. Most candidate virulence genes are not always connected with disease.[4]
The model of EAEC pathogenesis comprises three stages: Stage 1 is the attachment of the intestinal mucosa by aggregative adherence fimbriae and other adhering projections, Stage 2 an increase in mucus that covers EAEC on its surface of enterocytes is found; Stage 3 evocation of an inflammatory response, mucosal toxicity, and intestinal secretion as well as a release of toxins exist.[21][4]
Stage one: Aggregative adherence factors (AAF) are responsible for the adhesion to the intestinal mucosa. AAF are made up of three fimbriae encoded by the pAA plasmid; aag aafA agg-3. aggA is in charge of aggregative phenotype and human erythrocyte haemagglutination of EAEC. aafA allows EAEC to adhere to the intestinal mucosa. agg-3 serves as an adhesion. MAP, three-membrane associated proteins, are essential in the EAEC adherence to haemagglutination of animal cells.[4]
Stage two: After AAF factors in stage 1, adherence to the mucosa is characterized by the presence a biofilm. The production of biofilm is regulated by AggR and demands several genes. The loss of biofilm production and diffuse adherence pattern was reported in EAEC at a pH of 4.0. Many studies reveal that EAEC are capable of surviving in the mucus layer. This evidence can support why malnourished children who are infected with EAEC and live in poor conditions develop mucoid stools and prolonged diarrhea.[4]
Stage three: Cytotoxic effects are found in the release of toxins in EAEC as well as an elicitation of the inflammatory response, mucosal toxicity, and intestinal secretion. EAEC toxins are destructive to the intestinal villi and enterocytes. There are three toxins found in EAEC; plasmid encoded toxin (Pet), heat-stable toxin (EAST1), and Shigella enterotoxin 1 (ShET1).[4]
Diagnosis
Diagnosis of infectious diarrhea and identification of antimicrobial resistance is performed using a stool culture with subsequent antibiotic sensitivity testing. It requires a minimum of 2 days and maximum of several weeks to culture gastrointestinal pathogens. The sensitivity (true positive) and specificity (true negative) rates for stool culture vary by pathogen, although a number of human pathogens can not be cultured.[22][23][24]
Current point of care molecular diagnostic tests can identify EAEC and antimicrobial resistance in the identified strains much faster than culture and sensitivity testing; Microarray-based platforms can identify EAEC and AMR genes in two hours or less with high sensitivity and specificity, but the size of the test panel is limited. [25][26][27]
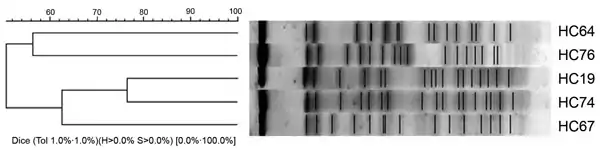 Cluster analysis of the enteroaggregative Escherichia coli strains from the pulsed-field gel electrophoresis fingerprinting.
Cluster analysis of the enteroaggregative Escherichia coli strains from the pulsed-field gel electrophoresis fingerprinting. Stool specimen container
Stool specimen container
Prevention
Proven prevention methods for E. coli transmission include handwashing and improved sanitation and drinking water, as transmission occurs through fecal contamination of food and water supplies. Additionally, thoroughly cooking meat and avoiding consumption of raw, unpasteurized beverages, such as juices and milk are other proven methods for preventing E. coli. Lastly, avoid cross-contamination of utensils and work spaces when preparing food.[28]
Treatment

Fluoroquinolone, especially ciprofloxacin, may be effective for Enteroaggregative E.coli (EAEC) infections, with a reductions in duration of diarrhea.[5][29]
For most people treatments include, rest and the intake of fluids. For patients with profuse diarrhea or vomit should be rehydrated by drinking much water or by drinking rehydration solutions such as Rehydralyte or Pedialyte.[30]
History

In 1885, the German-Austrian pediatrician Theodor Escherich discovered this organism in the feces of healthy individuals. He called it Bacterium coli commune because it is found in the colon. Early classifications of prokaryotes placed these in a handful of genera based on their shape and motility.[31][32][33]
Bacterium coli was the type species of the now invalid genus Bacterium when it was revealed that the former type species ("Bacterium triloculare") was missing.[34] Following a revision of Bacterium, it was reclassified as Bacillus coli by Migula in 1895[35] and later reclassified in the newly created genus Escherichia, named after its original discoverer, by Aldo Castellani and Albert John Chalmers.[36]
In 1996, the world's worst to date outbreak of E. coli food poisoning occurred in Wishaw, Scotland, killing 21 people.[37][38] This death toll was exceeded in 2011, when the 2011 Germany E. coli O104:H4 outbreak, linked to organic fenugreek sprouts, killed 53 people.[39]
Enteroaggregative Escheichia coli (EAEC) was first found in 1987, in a child in Lima, Peru.[7]
Since 1987, Enteroaggregative Escheichia coli (EAEC) has been recognized as agents of diarrhea in industrialized and developing countries, Enteroaggregative Escheichia coli is most commonly found in developing countries due to less developed industrial base and low human development compared to other countries (India, Jamaica and Mexico are the most commonly risked countries).[40][41]
Epidemiology
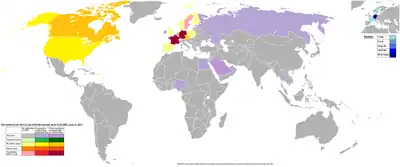
| Country | Non-HUS cases | HUS cases | Deaths |
|---|---|---|---|
| 4 | 1 | 0 | |
| 1 | 0 | 0 | |
| 1 | 0 | 0 | |
| 16 | 10 | 0 | |
| 4 | 9 | 0 | |
| 2947 | 818 | 51 | |
| 1 | 0 | 0 | |
| 1 | 1 | 0 | |
| 7 | 4 | 0 | |
| 1 | 0 | 0 | |
| 1 | 2 | 0 | |
| 1 | 1 | 0 | |
| 35 | 18 | 1 | |
| 5 | 0 | 0 | |
| 3 | 4 | 0 | |
| 2 | 4 | 1 | |
| Total | 2987 | 855 | 53 |
Escherichia coli O104:H4 is an enteroaggregative Escherichia coli strain of the bacterium Escherichia coli, and the cause of the 2011 Escherichia coli O104:H4 outbreak.[42]
Analysis of genomic sequences obtained by BGI Shenzhen shows that the O104:H4 outbreak strain is an enteroaggregative E. coli (EAEC or EAggEC) type that has acquired Shiga toxin genes, presumably by horizontal gene transfer.[43][44][45]
Cases began as early as 1 May 2011 with a man in Aachen reporting bloody diarrhea.[11]
Cases then rapidly increased, with over 100 cases of EHEC gastroenteritis and/or HUS were being reported each day by 16 May.[11]
The outbreak centered on the five northern German states of Hamburg, Schleswig-Holstein, Bremen, Lower Saxony, and Mecklenburg-Western Pomerania.[11]
Cases would eventually be reported in all 16 German states; however most cases outside of the northern states were linked to travel in northern Germany.[46]
Additionally, a small number of cases were reported from other countries, although most of those ill had previously travelled to Germany. The most substantial outbreak outside of Germany was in Bordeaux, France where 15 cases of EHEC gastroenteritis were associated with the same strain of E. coli which caused the outbreak in Germany. The French cases had not previously travelled to Germany, suggesting they acquired the bacteria from contaminated sprouts grown in France.[11]
Cases of EHEC HUS and gastroenteritis peaked on 21 and 22 May respectively.[46]
Cases then slowly decreased over the following month, with cases reported throughout the month of June and ending during July 2011. German authorities deemed the outbreak over in early July 2011.[11]
References
- 1 2 3 4 5 6 7 Jensen, Betina Hebbelstrup; Olsen, Katharina E. P.; Struve, Carsten; Krogfelt, Karen Angeliki; Petersen, Andreas Munk (2014). "Epidemiology and Clinical Manifestations of Enteroaggregative Escherichia coli". Clinical Microbiology Reviews. 27 (3): 614–630. doi:10.1128/CMR.00112-13. ISSN 0893-8512. PMC 4135892. PMID 24982324.
- 1 2 Nataro, J. P.; Mai, V.; Johnson, J.; Blackwelder, W. C.; Heimer, R.; Tirrell, S.; Edberg, S. C.; Braden, C. R.; Morris, J. G. (2006-08-15). "Diarrheagenic Escherichia coli Infection in Baltimore, Maryland, and New Haven, Connecticut". Clinical Infectious Diseases. 43 (4): 402–407. doi:10.1086/505867. ISSN 1058-4838. PMID 16838226. Archived from the original on 2017-02-08. Retrieved 2017-02-03.
- 1 2 Nataro, James P.; Steiner, Theodore (2002), "Enteroaggregative and Diffusely Adherent Escherichia Coli", Escherichia Coli, Elsevier, pp. 189–207, doi:10.1016/b978-012220751-8/50007-0, ISBN 9780122207518
- 1 2 3 4 5 6 7 Jenkins C (2018). "Enteroaggregative Escherichia coli". Current Topics in Microbiology and Immunology. 416: 27–50. doi:10.1007/82_2018_105. ISBN 978-3-319-99663-9. PMID 30232602.
- 1 2 3 Kaur, P.; Chakraborti, A.; Asea, A. (2010). "Enteroaggregative Escherichia coli : An Emerging Enteric Food Borne Pathogen". Interdisciplinary Perspectives on Infectious Diseases. 2010: 254159. doi:10.1155/2010/254159. ISSN 1687-708X. PMC 2837894. PMID 20300577.
- ↑ Hebbelstrup Jensen, Betina; Adler Sørensen, Camilla; Hebbelstrup Rye Rasmussen, Stig; Rejkjær Holm, Dorthe; Friis-Møller, Alice; Engberg, Jørgen; Mirsepasi-Lauridsen, Hengameh C.; Struve, Carsten; Hammerum, Anette M.; Porsbo, Lone Jannok; Petersen, Randi Føns; Petersen, Andreas Munk; Krogfelt, Karen Angeliki (2018). "Characterization of Diarrheagenic Enteroaggregative Escherichia coli in Danish Adults—Antibiotic Treatment Does Not Reduce Duration of Diarrhea". Frontiers in Cellular and Infection Microbiology. 8. ISSN 2235-2988. Archived from the original on 19 January 2023. Retrieved 18 September 2023.This, and other studies, have suggested that fluoroquinolones, especially ciprofloxacin, may be the most effective antibiotic when treating EAEC infections
- ↑ Huang, David B.; Mohanty, Alakananda; DuPont, Herbert L.; Okhuysen, Pablo C.; Chiang, Tom (2006-10-01). "A review of an emerging enteric pathogen: enteroaggregative Escherichia coli". Journal of Medical Microbiology. 55 (Pt 10): 1303–1311. doi:10.1099/jmm.0.46674-0. ISSN 0022-2615. PMID 17005776. S2CID 9536299.
- ↑ Adachi JA, Jiang ZD, Mathewson JJ, Verenkar MP, Thompson S, Martinez-Sandoval F, Steffen R, Ericsson CD, DuPont HL (2001). "Enteroaggregative Escherichia coli as a Major Etiologic Agent in Traveler's Diarrhea in 3 Regions of the World" (PDF). Clin Infect Dis. 32 (12): 1706–9. doi:10.1086/320756. PMID 11360211. Archived (PDF) from the original on 2017-02-03. Retrieved 2017-02-03.
- ↑ Huang, D. B.; et al. (2006). "A review of an emerging enteric pathogen: enteroaggregative Escherichia coli". Journal of Medical Microbiology. 55 (10): 1303–1311. doi:10.1099/jmm.0.46674-0. PMID 17005776. S2CID 9536299.
- 1 2 3 4 5 6 7 "Final presentation and evaluation of epidemiological findings in the EHEC O104:H4 outbreak – Germany 2011" (PDF). Robert Koch Institut. 2011. Archived from the original on 16 June 2019. Retrieved 12 January 2018.
- ↑ Kalita, Anjana; Hu, Jia; Torres, Alfredo G. (2014). "Recent advances in adherence and invasion of pathogenic Escherichia coli". Current Opinion in Infectious Diseases. 27 (5): 459–464. doi:10.1097/QCO.0000000000000092. ISSN 0951-7375. PMC 4169667. PMID 25023740.
- ↑ Nadia Boisen; Angela R. Melton-Celsa; Flemming Scheutz; Alison D. O'Brien; James P. Nataro (2015). "Shiga toxin 2a and Enteroaggregative Escherichia coli—a deadly combination". Gut Microbes. 6 (4): 272–278. doi:10.1080/19490976.2015.1054591. PMC 4615819. PMID 26039753.
- ↑ "Outbreak of Escherichia coli O104:H4 Infections Associated with Sprout Consumption — Europe and North America, May–July 2011". www.cdc.gov. Archived from the original on 2017-01-18. Retrieved 2017-02-01.
- ↑ Ruan, Xiaosai; Crupper, Scott S.; Schultz, Bruce D.; Robertson, Donald C.; Zhang, Weiping (2012-08-15). "Escherichia coli Expressing EAST1 Toxin Did Not Cause an Increase of cAMP or cGMP Levels in Cells, and No Diarrhea in 5-Day-Old Gnotobiotic Pigs". PLOS ONE. 7 (8): e43203. Bibcode:2012PLoSO...743203R. doi:10.1371/journal.pone.0043203. ISSN 1932-6203. PMC 3419656. PMID 22905235.
- ↑ "Questions and Answers | E. coli | CDC". www.cdc.gov. 11 January 2019. Archived from the original on 17 May 2017. Retrieved 26 August 2022.
- ↑ "What are the incubation periods for infections?". 2018-06-26. Archived from the original on 2019-11-10.
- ↑ Kampmeier S, Berger M, Mellmann A, Karch H, Berger P (2018). "The 2011 German Enterohemorrhagic Escherichia Coli O104:H4 Outbreak-The Danger Is Still Out There". Current Topics in Microbiology and Immunology. 416: 117–148. doi:10.1007/82_2018_107. ISBN 978-3-319-99663-9. PMID 30062592.
- 1 2 Hebbelstrup Jensen, Betina; Olsen, Katharina E. P.; Struve, Carsten; Krogfelt, Karen Angeliki; Petersen, Andreas Munk (July 2014). "Epidemiology and Clinical Manifestations of Enteroaggregative Escherichia coli". Clinical Microbiology Reviews. 27 (3): 614–630. doi:10.1128/CMR.00112-13. Archived from the original on 25 April 2022. Retrieved 27 August 2022.
- ↑ Oundo, Joseph O.; Kariuki, Samuel M.; Boga, Hamadi I.; Muli, Faith W.; Iijima, Yoshio (January 2008). "High incidence of enteroaggregative Escherichia coli among food handlers in three areas of Kenya: a possible transmission route of travelers' diarrhea". Journal of Travel Medicine. 15 (1): 31–38. doi:10.1111/j.1708-8305.2007.00174.x. ISSN 1195-1982. Archived from the original on 7 February 2022. Retrieved 27 August 2022.
- ↑ Nataro, James P. (January 2005). "Enteroaggregative Escherichia coli pathogenesis". Current Opinion in Gastroenterology. 21 (1): 4–8. ISSN 0267-1379. Archived from the original on 21 May 2017. Retrieved 25 August 2022.
- ↑ Hewison, Christopher J.; Heath, Christopher H.; Ingram, Paul R. (October 2012). "Stool culture". Australian Family Physician. 41 (10): 775–779. ISSN 0300-8495. PMID 23210099. Archived from the original on 13 September 2022. Retrieved 1 September 2022.
- ↑ "Antibiotic Sensitivity Test: MedlinePlus Medical Test". medlineplus.gov. Archived from the original on 4 August 2022. Retrieved 24 August 2022.
- ↑ Nataro, James (June 1998). "Enteroaggregative Escherichia coli". Emerging Infectious Diseases. 4 (2): 251–261. doi:10.3201/eid0402.980212. Archived from the original on 13 September 2022. Retrieved 28 August 2022.
- ↑ "Diagnostic Methods for the Enteroaggregative Escherichia coli Infection". American Society of Microbiology. Archived from the original on 25 August 2022. Retrieved 23 August 2022.
- ↑ Law, D.; Chart, H. (May 1998). "Enteroaggregative Escherichia coli". Journal of Applied Microbiology. 84 (5): 685–697. doi:10.1046/j.1365-2672.1998.00372.x. ISSN 1364-5072. Archived from the original on 7 February 2022. Retrieved 23 August 2022.
- ↑ Sahadulla, M. I.; Uduman, Sayenna A. (31 July 2019). Comprehensive Textbook of Infectious Diseases. Jaypee Brothers Medical Publishers. p. 205. ISBN 978-93-5270-999-1. Archived from the original on 13 September 2022. Retrieved 2 September 2022.
- ↑ "General Information- E.coli". Centers for Disease Control and Prevention. Archived from the original on 17 May 2017. Retrieved 25 May 2017.
- ↑ Bobat, Raziya (27 February 2020). HIV Infection in Children and Adolescents. Springer Nature. p. 81. ISBN 978-3-030-35433-6. Archived from the original on 25 August 2022. Retrieved 24 August 2022.
- ↑ "E. coli – treatments and diagnosis". Mayo Clinic. Archived from the original on 2019-11-12. Retrieved 2019-11-12.
- ↑ Farrar J, Hotez P, Junghanss T, Kang G, Lalloo D, White NJ, eds. (2013). Manson's Tropical Diseases (23rd ed.). Oxford: Elsevier/Saunders. ISBN 978-0702053061.
- ↑ Haeckel E (1867). Generelle Morphologie der Organismen. Reimer, Berlin. ISBN 978-1-144-00186-3.
- ↑ Escherich T (1885). "Die Darmbakterien des Neugeborenen und Säuglinge". Fortschr. Med. 3: 515–22. Archived from the original on 2021-08-17. Retrieved 2022-08-21.
- ↑ Breed RS, Conn HJ (May 1936). "The Status of the Generic Term Bacterium Ehrenberg 1828". Journal of Bacteriology. 31 (5): 517–18. doi:10.1128/JB.31.5.517-518.1936. PMC 543738. PMID 16559906.
- ↑ Migula W (1895). "Bacteriaceae (Stabchenbacterien)". In Engerl A, Prantl K (eds.). Die Naturlichen Pfanzenfamilien, W. Engelmann, Leipzig, Teil I, Abteilung Ia. pp. 20–30.
- ↑ Castellani A, Chalmers AJ (1919). Manual of Tropical Medicine (3rd ed.). New York: Williams Wood and Co.
- ↑ "Sheriff criticises E. Coli butcher". BBC News. Archived from the original on 2007-01-08. Retrieved 2022-08-21.
- ↑ "The butcher who lied". HeraldScotland. Archived from the original on 2021-10-28. Retrieved 2021-10-15.
- ↑ European Food Safety Authority (11 July 2012). "E.coli: Rapid response in a crisis". Archived from the original on 20 November 2018. Retrieved 2 October 2012.
there were 53 confirmed deaths.
- ↑ "Enteroaggregative Escherichia coli" (PDF). CDC. Archived (PDF) from the original on 3 April 2021. Retrieved 26 August 2022.
- ↑ Cennimo, David J.; Koo, Hoonmo; Mohamed, Jamal A.; Huang, David B.; Chiang, Tom (March 2007). "Enteroaggregative Escherichia coli: A review of trends, diagnosis, and treatment". Infections in Medicine. 24 (3): 100–110. ISSN 0749-6524. Archived from the original on 13 September 2022. Retrieved 31 August 2022.
- ↑ Mellman, Alexander; Harmsen, D; Cummings, CA; et al. (July 20, 2011). "Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology". PLoS One. 6 (7): e22751. doi:10.1371/journal.pone.0022751. PMC 3140518. PMID 21799941.
- ↑ "BGI Sequences Genome of the Deadly E. coli in Germany and Reveals New Super-Toxic Strain". BGI. 2011-06-02. Archived from the original on 2011-06-06. Retrieved 2011-06-02.
- ↑ David Tribe (2011-06-02). "BGI Sequencing news: German EHEC strain is a chimera created by horizontal gene transfer". Biology Fortified. Archived from the original on 2012-05-27. Retrieved 2011-06-02.
- ↑ Maev Kennedy and agencies (2011-06-02). "E. coli outbreak: WHO says bacterium is a new strain". London: guardian.co.uk. Archived from the original on 2011-06-02. Retrieved 2011-06-04.
- 1 2 Frank, Christina; Werber, Dirk; Cramer, Jakob P.; Askar, Mona; Faber, Mirko; An Der Heiden, Matthias; Bernard, Helen; Fruth, Angelika; Prager, Rita; Spode, Anke; Wadl, Maria; Zoufaly, Alexander; Jordan, Sabine; Kemper, Markus J.; Follin, Per; Müller, Luise; King, Lisa A.; Rosner, Bettina; Buchholz, Udo; Stark, Klaus; Krause, Gérard; HUS Investigation Team (2011). "Epidemic Profile of Shiga-Toxin–Producing Escherichia coliO104:H4 Outbreak in Germany". New England Journal of Medicine. 365 (19): 1771–1780. doi:10.1056/NEJMoa1106483. PMID 21696328.