Fis
| DNA-binding protein Fis | |||||||
|---|---|---|---|---|---|---|---|
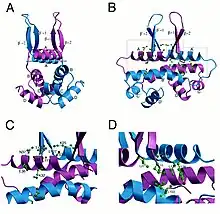 Fis K36E homodimer structure | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | fis | ||||||
| UniProt | P0A6R3 | ||||||
| |||||||
fis is an E. coli gene encoding the Fis (or FIS) protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.
History
Fis was first discovered for its role in stimulating Gin catalyzed inversion of the G segment of phage Mu genome.[1] Fis was originally identified as the factor for inversion stimulation of the homologous Hin and Gin site-specific DNA recombinases of Salmonella and phage Mu, respectively. This small, basic, DNA-bending protein has recently been shown to function in many other reactions including phage lambda site-specific recombination, transcriptional activation of rRNA and tRNA operons, repression of its own synthesis, and oriC-directed DNA replication. Cellular concentrations of Fis vary tremendously under different growth conditions which may have important regulatory implications for the physiological role of Fis in these different reactions.[2]
Structure
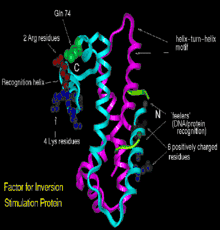
Structurally, Fis folds into four α-helices (A–D) and a β-hairpin. Helices A and B provide the contacts between Fis monomers, facilitating dimer formation, whereas the C and D helices form a helix-turn-helix motif that is essential for DNA binding.[3]
Properties and functions
Fis is a very important small nucleotide-associated protein which plays a role in affecting the bacterial chromosome structure and the initiation of DNA replication.[4] It is a nucleoid-associated protein in Escherichia coli that is abundant during early exponential growth in rich medium but is in short supply during stationary phase.[5] When stationary-phase cells are subcultured into a rich medium, Fis levels increase from less than 100 to over 50,000 copies per cell prior to the first cell division. As cells enter exponential growth, nascent synthesis is largely shut off, and intracellular Fis levels decrease as a function of cell division. Fis synthesis also transiently increases when exponentially growing cells are shifted to a richer medium. The magnitude of the peak of Fis synthesis appears to reflect the extent of the nutritional upshift. fis mRNA levels closely resemble the protein expression pattern, suggesting that regulation occurs largely at the transcriptional level. Two RNA polymerase-binding sites and at least six high-affinity Fis-binding sites are present in the fis promoter region. Expression of this fis operon is negatively regulated by Fis in vivo and purified Fis can prevent stable complex formation by RNA polymerase at the fis promoter in vitro. However, autoregulation only partially accounts for the expression pattern of Fis. Fluctuations in Fis levels have been shown to serve as an early signal of a nutritional upshift and is important in the physiological roles Fis plays in the cell.[6]
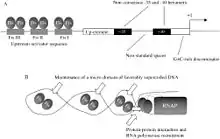
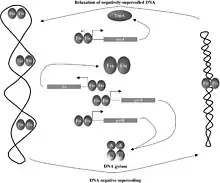
It is a global regulatory protein in Escherichia coli that activates ribosomal RNA (rRNA) transcription by binding to three upstream activation sites of the rRNA promoter and enhances transcription 5 to 10 fold in vivo. Fis overexpression results in different effects on cell growth depending on nutrient conditions.[7] The Fis nucleoid protein is differentiated by its fast increase in synthesis rates following nutrient upshifts and its abundance in rapidly growing E. coli cells.[8]
Fis has been known to activate ribosomal RNA transcription, as well other genes. It has a direct role in upstream activation of rRNA promoters. Fis binds to a recombinational enhancer sequence that is required to stimulate hin-mediated DNA inversion. It has also been shown to prevent initiation of DNA replication from oriC.[9]
It has been shown that sequences from 32 to 94 nucleotides upstream of the fis AUG are responsible for increasing fis lacZ translation reporter activities over 100 fold. Within this region, an AU sequence element centered 35 nucleotides upstream of the fis AUG increases fis translation by as much as 15 fold. Formation of a supposed RNA secondary structure element beginning 50 nucleotides upstream of the AUG also positively affects fis translation by up to 10 fold. The fis gene is cotranscribed with the upstream dusB gene encoding a tRNA-modifying enzyme. DusB protein levels are very low even under conditions when there is high transcription of the operon and high levels of Fis protein.[10]
Fis has been deemed a bacterial chromatin architectural protein.[11] Besides modulating chromatin architecture, it is known to influence numerous promoters of E. coli and several other bacteria. Both in vivo and in vitro studies indicate that Fis acts as a transcriptional repressor of ''mom'' promoter. There is data that shows Fis mediates its repressive effect by denying access to RNA polymerase at the mom promoter. combined A repressive effect of Fis and previously characterized negative regulatory factors could be responsible to keep the gene silenced most of the time. In addition to bringing about overall downregulation of the Mu genome, it also ensures silencing of the advantageous but potentially lethal mom gene.[12]
Fis as a critical regulator of capsule expression. Fis is also involved in the regulation of a range of genes in bacterial species such as P. multocida, Enteroaggregative Escherichia coli,[13] similar organisms. Some of these genes include important virulence factors.[14]
Role of Fis in bacterial motility
The role of fis is well studied in E. coli, but its role in pseudomonads has only been examined briefly. Recent studies in Enterobacteriaceae have shown that fis positively regulates the flagellar movement of bacteria. Observations in Pseudomonas putida demonstrate fis reduced the migration of P. putida onto the apices of barley roots and thereby the competitiveness of bacteria on the roots. It was also observed that the overexpression of fis drastically reducing swimming motility and facilitated the formation of P. putida biofilm. It is possible that the elevated expression of Fis is important in the adaptation of P. putida during colonization of plant roots by promoting biofilm formation when the migration of bacteria is no longer advantageous.[15]
It was demonstrated that Fis is essential for the stability of the linear plasmid pDSIUDi and affects the motility of S. Typhi.[16]
Fis buffers decrease of negative supercoiling in tyrT and rrnA expression. The upstream FIS binding site of rrnA is required for this and it's probable that FIS enables local DNA curvature. See Travers and Muskhelishvili 2005 for more detail.
References
- ↑ Koch C., Kahmann R. (1986). "Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu". J. Biol. Chem. 261 (33): 15673–15678. doi:10.1016/S0021-9258(18)66770-5. PMID 3536909.
- ↑ Finkel, S. E., and R. C. Johnson. "The Fis protein: it's not just for DNA inversion anymore." Molecular microbiology 6.22 (2006): 3257-3265.
- ↑ Steen JA, Steen JA, Harrison P, Seemann T, Wilkie I; et al. (2010). "Fis Is Essential for Capsule Production in Pasteurella multocida and Regulates Expression of Other Important Virulence Factors". PLOS Pathog. 6 (2): e1000750. doi:10.1371/journal.ppat.1000750. PMC 2816674. PMID 20140235.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Wold S, Crooke E, Skarstad K (1996). "The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro". Nucleic Acids Res. 24 (18): 3527–3532. doi:10.1093/nar/24.18.3527. PMC 146119. PMID 8836178.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Bradley Meranda D; et al. (2007). "Effects of Fis on Escherichia coli gene expression during different growth stages". Microbiology. 153 (9): 2922–2940. doi:10.1099/mic.0.2007/008565-0. PMID 17768236.
- ↑ Ball Catherine A; et al. (1992). "Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli". Journal of Bacteriology. 174 (24): 8043–8056. doi:10.1128/jb.174.24.8043-8056.1992. PMC 207543. PMID 1459953.
- ↑ Leila H. Choe, Wilfred Chen, Kelvin H. Lee, Proteome analysis of factor for inversion stimulation (Fis) overproduction in Escherichia coli,Electrophoresis 1999, 20, 798-805
- ↑ Nafissi Maryam; et al. (2012). "Robust translation of the nucleoid protein Fis requires a remote upstream AU element and is enhanced by RNA secondary structure". Journal of Bacteriology. 194 (10): 2458–2469. doi:10.1128/jb.00053-12. PMC 3347164. PMID 22389479.
- ↑ Ross W., Thompson J.F., Newlands J.T., Gourse R.L (1990). "E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo". EMBO J. 9 (11): 3733–3742. doi:10.1002/j.1460-2075.1990.tb07586.x. PMC 552129. PMID 2209559.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Nafissi Maryam; et al. (2012). "Robust translation of the nucleoid protein Fis requires a remote upstream AU element and is enhanced by RNA secondary structure". Journal of Bacteriology. 194 (10): 2458–2469. doi:10.1128/jb.00053-12. PMC 3347164. PMID 22389479.
- ↑ Travers, A.; Schneider, R.; Muskhelishvili, G. (2001). "DNA supercoiling and transcription in Escherichia coli: The FIS connection". Biochimie. 83 (2): 213–7. doi:10.1016/S0300-9084(00)01217-7. PMID 11278071.
- ↑ Karambelkar Shweta, Swapna Ganduri, Nagaraja Valakunja (2012). "Silencing of toxic gene expression by Fis". Nucleic Acids Research. 40 (10): 4358–4367. doi:10.1093/nar/gks037. PMC 3378877. PMID 22287621.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Rossiter, Amanda E., et al. "Transcription of the plasmid‐encoded toxin gene from Enteroaggregative Escherichia coli is regulated by a novel co‐activation mechanism involving CRP and Fis." Molecular microbiology 81.1 (2011): 179-191.
- ↑ Steen JA, Steen JA, Harrison P, Seemann T, Wilkie I; et al. (2010). "Fis Is Essential for Capsule Production in Pasteurella multocida and Regulates Expression of Other Important Virulence Factors". PLOS Pathog. 6 (2): e1000750. doi:10.1371/journal.ppat.1000750. PMC 2816674. PMID 20140235.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Jakovleva Julia; et al. (2012). "Fis regulates the competitiveness of Pseudomonas putida on barley roots by inducing biofilm formation". Microbiology. 158 (3): 708–720. doi:10.1099/mic.0.053355-0. PMID 22222498.
- ↑ Zhang Haifang; et al. (2012). "Fis Is Essential for the Stability of Linear Plasmid pBSSB1 and Affects the Motility of Salmonella enterica Serovar Typhi". PLOS ONE. 7 (7): e37462. Bibcode:2012PLoSO...737462Z. doi:10.1371/journal.pone.0037462. PMC 3402438. PMID 22911678.