Paramyxoviridae
{{ |image =12985 2019 1136 Fig1 HTML.webp |image_alt = |image_caption =Canine distemper virus (CDV) virion and genome organization |taxon =Paramyxoviridae |subdivision_ranks=Subfamilies |subdivision =* Avulavirinae
- Metaparamyxovirinae
- Orthoparamyxovirinae
- Rubulavirinae
}} Paramyxoviridae (from Greek para- “by the side of” and myxa “mucus”) is a family of negative-strand RNA viruses in the order Mononegavirales.[1][2] Vertebrates serve as natural hosts.[3] Diseases associated with this family include measles, mumps, and respiratory tract infections.[4] The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.[5]
Structure
Virions are enveloped and can be spherical or pleomorphic and capable of producing filamentous virions. The diameter is around 150 nm. Genomes are linear, around 15kb in length.[6][1] Fusion proteins and attachment proteins appear as spikes on the virion surface. Matrix proteins inside the envelope stabilise virus structure. The nucleocapsid core is composed of the genomic RNA, nucleocapsid proteins, phosphoproteins and polymerase proteins.[7][8]
Genome
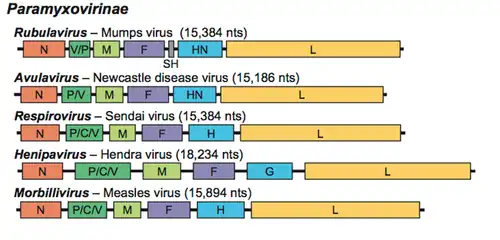
The genome is nonsegmented, negative-sense RNA, 15–19 kilobases in length, and contains six to 10 genes. Extracistronic (noncoding) regions include:[9][10][11]
- A 3’ leader sequence, 50 nucleotides in length, which acts as a transcriptional promoter.
- A 5’ trailer sequence, 50–161 nucleotides long
- Intergenomic regions between each gene, which are three nucleotides long for morbilliviruses, respiroviruses, and henipaviruses, and variable length for rubulaviruses.
Each gene contains transcription start/stop signals at the beginning and end, which are transcribed as part of the gene.The gene sequence within the genome is conserved across the family due to a phenomenon known as transcriptional polarity in which genes closest to the 3’ end of the genome are transcribed in greater abundance than those towards the 5’ end, this is a result of structure of the genome. After each gene is transcribed, the RNA-dependent RNA polymerase pauses to release the new mRNA when it encounters an intergenic sequence. When the RNA polymerase is paused, a chance exists that it will dissociate from the RNA genome. If it dissociates, it must re-enter the genome at the leader sequence, rather than continuing to transcribe the length of the genome.[9][10][12][13]
Single promoter model was verified when viruses were exposed to UV light, UV radiation can cause dimerization of RNA, which prevents transcription by RNA polymerase. When paramyxovirus genome was exposed to UV light, the level of inhibition of transcription was proportional to the distance from the leader sequence, in other words, the further the gene is from the leader sequence, the greater the chance of RNA dimerization inhibiting RNA polymerase.The virus takes advantage of the single promoter model by having its genes arranged in relative order of protein needed for successful infection.[10][14][15][16]
Viruses in the Paramyxoviridae family are also antigenically stable[17], meaning that the glycoproteins on the viruses are consistent between different strains of the same type, two reasons for this phenomenon are posited:
- The first is that the genome is nonsegmented, thus cannot undergo genetic reassortment. For this process to occur, segments needed as reassortment happen when segments from different strains are mixed together to create a new strain.hence with no segments, nothing can be mixed with one another, so no antigenic shift occurs.[18][19]
- The second reason relates to the idea of genetic variation, since RNA-dependent RNA polymerase does not have an error-checking function, many mutations are made when the RNA is processed, these mutations build up and eventually new strains are created. Due to this, one would expect that paramyxoviruses should not be antigenically stable; however, it is. The main hypothesis behind why the viruses are antigenically stable is that each protein and amino acid has an important function. Thus, any mutation would lead to a decrease or total loss of function, which would in turn cause the new virus to be less efficient. These viruses would not be able to survive as long compared to the more virulent strains, and so would die out.Many paramyxovirus genomes follow the "rule of six", the total length of the genome is almost always a multiple of six.[20][14][21]
Proteins
Paramyxoviridae virion is as follows:
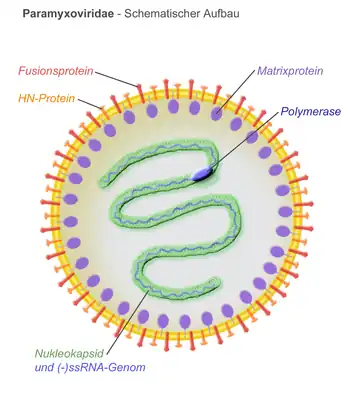
- N – the nucleocapsid protein associates with genomic RNA (one molecule per hexamer) and protects the RNA from nuclease digestion[22]
- P – the phosphoprotein binds to the N and L proteins and forms part of the RNA polymerase complex. P is the polymerase co-factor.[23]
- M – the matrix protein assembles between the envelope and the nucleocapsid core, it organizes and maintains virion structure[8]
- F – the fusion protein projects from the envelope surface as a trimer, and mediates cell entry by inducing fusion between the viral envelope and the cell membrane by class I fusion. One of the defining characteristics of members of the family Paramyxoviridae is the requirement for a neutral pH for fusogenic activity.[24]
- H/HN/G – the cell attachment proteins span the viral envelope and project from the surface as spikes. They bind to proteins on the surface of target cells to facilitate cell entry. Proteins are designated H for morbilliviruses as they possess haemagglutination activity, observed as an ability to cause red blood cells to clump in laboratory tests. HN attachment proteins occur in respiroviruses, rubulaviruses and avulaviruses. These possess both haemagglutination and neuraminidase activity, which cleaves sialic acid on the cell surface, preventing viral particles from reattaching to previously infected cells. Attachment proteins with neither haemagglutination nor neuraminidase activity are designated G. These occur in henipaviruses.[25][15][26]
- L – the large protein is the catalytic subunit of RNA-dependent RNA polymerase (RDRP)[27]
- Accessory proteins – a mechanism known as RNA editing allows multiple proteins to be produced from the P gene. These are not essential for replication but may aid in survival in vitro or may be involved in regulating the switch from mRNA synthesis to antigenome synthesis.[28][29]
Life cycle
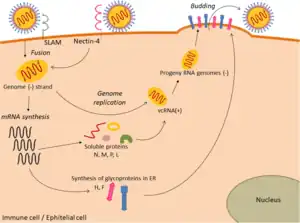
Viral replication is cytoplasmic. Entry into the host cell is achieved by viral attachment to host cell. Replication and transcription follow the negative-stranded RNA virus models.[30] Translation takes place by leaky scanning, ribosomal shunting, and RNA termination-reinitiation. The virus exits the host cell by budding. Human, vertebrates, and birds serve as the natural hosts. Transmission route is airborne particles.[1]
The Paramyxoviridae are able to undergo mRNA editing, which produces different proteins from the same mRNA transcript by slipping back one base to read off in a different open reading frame (ORF) due to the presence of secondary structures such as pseudoknots. Paramyxoviridae also undergo transcriptional stuttering to produce the poly (A) tail at the end of mRNA transcripts by repeatedly moving back one nucleotide at a time at the end of the RNA template.[31][32]
Taxonomy

Family: Paramyxoviridae[5]
- Subfamily: Avulavirinae, which contains three genera and 22 species
- Subfamily: Metaparamyxovirinae, which contains one genus and one species
- Subfamily: Orthoparamyxovirinae, which contains eight genera and 34 species
- Subfamily: Rubulavirinae, which contains two genera and 18 species
- Unassigned genera:
- Cynoglossusvirus
- Hoplichthysvirus
- Scoliodonvirus
Pathogenic paramyxoviruses

A number of important human diseases are caused by paramyxoviruses. These include mumps, as well as measles, which caused around 733,000 deaths in 2000.[34]
The human parainfluenza viruses (HPIV) are the second most common causes of respiratory tract disease in infants and children. There are four types of HPIVs, known as HPIV-1, HPIV-2, HPIV-3 and HPIV-4. HPIV-1 and HPIV-2 may cause cold-like symptoms, along with croup in children. HPIV-3 is associated with bronchiolitis, bronchitis, and pneumonia. HPIV-4 is less common than the other types, and is known to cause mild to severe respiratory tract illnesses.[35]
Measles
Measles is a highly contagious infectious disease caused by measles virus.[36][37]
Symptoms usually develop 10–12 days after exposure to an infected person and last 7–10 days.[38][39] Initial symptoms typically include fever, often greater than 40 °C (104 °F), cough, runny nose, and inflamed eyes.[36][40] Small white spots known as Koplik's spots may form inside the mouth two or three days after the start of symptoms.[40] A red, flat rash which usually starts on the face and then spreads to the rest of the body typically begins three to five days after the start of symptoms.[40]
Mumps
Mumps is a viral disease caused by the mumps virus.[41] Initial signs and symptoms often include fever, muscle pain, headache, poor appetite, and feeling generally unwell.[42] This is then usually followed by painful swelling of one or both parotid salivary glands.[43][42]
Human parainfluenza viruses
It is estimated that there are 5 million children with lower respiratory infections (LRI) each year in the United States alone.[44] HPIV-1, HPIV-2 and HPIV-3 have been linked with up to a third of these infections.[45] Upper respiratory infections (URI) are also important in the context of HPIV, however, they are caused to a lesser extent by the virus.[46]
Other animals
Paramyxoviruses are also responsible for a range of diseases in other animal species, for example canine distemper virus (dogs), phocine distemper virus (seals), Newcastle disease virus (birds), and rinderpest virus (cattle). [47][48][49][50]
Some paramyxoviruses, such as the henipaviruses, are zoonotic pathogens, occurring naturally in an animal host, but also able to infect humans.[51]
Hendra virus (HeV) and Nipah virus (NiV) in the genus Henipavirus have emerged in humans and livestock in Australia and Southeast Asia. Both viruses are contagious, highly virulent, and capable of infecting a number of mammalian species and causing potentially fatal disease. Due to the lack of a licensed vaccine or antiviral therapies, HeV and NiV are designated as Biosafety level (BSL) 4 agents. The genomic structure of both viruses is that of a typical paramyxovirus.[52]
Diversity and evolution

In the past few decades, paramyxoviruses have been discovered from terrestrial, volant and aquatic animals, demonstrating a vast host range and great viral genetic diversity.[53]
As molecular technology advances and viral surveillance programs are implemented, the discovery of new viruses in this group is increasing.The evolution of paramyxoviruses is still debated, as McCarthy et al., indicates "the evolution of paramyxoviruses is still poorly understood". Using pneumoviruses as an outgroup, paramyxoviruses can be divided into two clades: one consisting of avulaviruses and rubulaviruses and one consisting of respiroviruses, henipaviruses, and morbilliviruses. Within the second clade the respiroviruses appear to be the basal group. The respirovirus-henipavirus-morbillivirus clade may be basal to the avulavirus-rubulavirus clade.[54][4][55][56]
See also
- Animal virology
- Virology
References
- 1 2 3 "Viral Zone". ExPASy. Archived from the original on 29 April 2017. Retrieved 15 June 2015.
- ↑ "Paramyxoviridae - Paramyxoviridae - Negative-sense RNA Viruses - ICTV". talk.ictvonline.org. Archived from the original on 2022-07-11. Retrieved 2020-12-14.
- ↑ Fields virology. Fields, Bernard N., Knipe, David M. (David Mahan), 1950-, Howley, Peter M. (6th ed.). Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins. 2013. p. 883. ISBN 9781451105636. OCLC 825740706.
{{cite book}}: CS1 maint: others (link) - 1 2 Samal, SK, ed. (2011). The Biology of Paramyxoviruses. Caister Academic Press. ISBN 978-1-904455-85-1.
- 1 2 "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Archived from the original on 20 March 2020. Retrieved 18 May 2021.
- ↑ Rima, B; Balkema-Buschmann, A; Dundon, WG; Duprex, WP; Easton, A; Fouchier, R; Kurath, G; Lamb, R; Lee, B; Rota, P; Wang, L; ICTV Report Consortium (December 2019). "ICTV Virus Taxonomy Profile: Paramyxoviridae". The Journal of General Virology. 100 (12): 1593–1594. doi:10.1099/jgv.0.001328. PMC 7273325. PMID 31609197.
- ↑ Morrison, Trudy G. (1 January 1998). "Paramyxoviruses, Infection and Immunity". Encyclopedia of Immunology (Second Edition). Elsevier. pp. 1909–1916. ISBN 978-0-12-226765-9. Retrieved 20 March 2023.
- 1 2 Cox, Robert M; Plemper, Richard K (2017). "Structure and Organization of Paramyxovirus Particles". Current opinion in virology. 24: 105–114. doi:10.1016/j.coviro.2017.05.004. ISSN 1879-6257. Archived from the original on 26 March 2023. Retrieved 26 March 2023.
- 1 2 Mishra, Pushpendra Mani; Verma, Navneet Chandra; Rao, Chethana; Uversky, Vladimir N.; Nandi, Chayan Kanti (1 January 2020). "Chapter One - Intrinsically disordered proteins of viruses: Involvement in the mechanism of cell regulation and pathogenesis". Progress in Molecular Biology and Translational Science. Academic Press. pp. 1–78. Archived from the original on 11 October 2022. Retrieved 20 March 2023.
- 1 2 3 Noton, Sarah L.; Fearns, Rachel (May 2015). "Initiation and regulation of paramyxovirus transcription and replication". Virology. 479–480: 545–554. doi:10.1016/j.virol.2015.01.014. ISSN 1096-0341. Archived from the original on 21 October 2022. Retrieved 22 March 2023.
- ↑ "Family: Paramyxoviridae | ICTV". ictv.global. Archived from the original on 11 July 2022. Retrieved 3 April 2023.
- ↑ Cox, Robert; Plemper, Richard K. (12 May 2015). "The paramyxovirus polymerase complex as a target for next-generation anti-paramyxovirus therapeutics". Frontiers in Microbiology. 6: 459. doi:10.3389/fmicb.2015.00459. ISSN 1664-302X. Archived from the original on 21 April 2022. Retrieved 1 April 2023.
- ↑ Kuo, L; Fearns, R; Collins, P L (September 1996). "The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome". Journal of Virology. 70 (9): 6143–6150. doi:10.1128/jvi.70.9.6143-6150.1996. ISSN 0022-538X.
- 1 2 Knipe, David Mahan; Howley, Peter M. (2007). Fields' Virology. Lippincott Williams & Wilkins. p. 1451. ISBN 978-0-7817-6060-7. Archived from the original on 24 March 2023. Retrieved 24 March 2023.
- 1 2 Enders, Gisela (1996). "Paramyxoviruses". Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. Archived from the original on 2023-03-06. Retrieved 2 April 2023.
- ↑ Acheson, N (2011). Fundamentals of Molecular Virology. Wiley. p. 181. ISBN 9780470900598. Archived from the original on 3 April 2023. Retrieved 3 April 2023.
- ↑ Ananthanarayan, R. (2006). Ananthanarayan and Paniker's Textbook of Microbiology. Orient Blackswan. p. 512. ISBN 978-81-250-2808-6. Archived from the original on 30 March 2023. Retrieved 30 March 2023.
- ↑ Schneider-Schaulies, Sibylle; Meulen, Volker ter (1 January 1999). "MEASLES VIRUS (PARAMYXOVIRIDAE)". Encyclopedia of Virology (Second Edition). Elsevier. pp. 952–960. ISBN 978-0-12-227030-7. Archived from the original on 20 October 2021. Retrieved 30 March 2023.
- ↑ Delost, Maria Dannessa (15 December 2020). Introduction to Diagnostic Microbiology for the Laboratory Sciences. Jones & Bartlett Learning. p. 502. ISBN 978-1-284-19973-4. Archived from the original on 2 April 2023. Retrieved 2 April 2023.
- ↑ Easton, AJ and Ling R (2010). Mahy BWJ and Van Regenmortel MHV (ed.). Desk Encyclopedia of General Virology. Academic Press. p. 496. ISBN 9780123751461..
- ↑ Kolakofsky, Daniel (1 November 2016). "Paramyxovirus RNA synthesis, mRNA editing, and genome hexamer phase: A review". Virology. 498: 94–98. doi:10.1016/j.virol.2016.08.018. ISSN 0042-6822. Retrieved 4 April 2023.
- ↑ Bloyet, Louis-Marie (9 December 2021). "The Nucleocapsid of Paramyxoviruses: Structure and Function of an Encapsidated Template". Viruses. 13 (12): 2465. doi:10.3390/v13122465. ISSN 1999-4915. Archived from the original on 16 November 2022. Retrieved 26 March 2023.
- ↑ Fuentes, Sandra M.; Sun, Dengyun; Schmitt, Anthony P.; He, Biao (January 2010). "Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression". Future Microbiology. 5 (1): 9–13. doi:10.2217/fmb.09.93. ISSN 1746-0921. Archived from the original on 26 March 2023. Retrieved 26 March 2023.
- ↑ Morrison, Trudy G. (11 July 2003). "Structure and function of a paramyxovirus fusion protein". Biochimica Et Biophysica Acta. 1614 (1): 73–84. doi:10.1016/s0005-2736(03)00164-0. ISSN 0006-3002. Archived from the original on 15 June 2022. Retrieved 26 March 2023.
- ↑ Bossart, Katharine N.; Fusco, Deborah L.; Broder, Christopher C. (2013). Paramyxovirus Entry. Landes Bioscience. Archived from the original on 26 March 2023. Retrieved 26 March 2023.
- ↑ Jardetzky, Theodore S.; Lamb, Robert A. (April 2014). "Activation of paramyxovirus membrane fusion and virus entry". Current Opinion in Virology. 5: 24–33. doi:10.1016/j.coviro.2014.01.005. ISSN 1879-6265. Archived from the original on 29 January 2023. Retrieved 30 March 2023.
- ↑ Venkataraman, Sangita; Prasad, Burra V. L. S.; Selvarajan, Ramasamy (10 February 2018). "RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution". Viruses. 10 (2): 76. doi:10.3390/v10020076. ISSN 1999-4915. Archived from the original on 30 January 2023. Retrieved 27 March 2023.
- ↑ Gotoh, B.; Komatsu, T.; Takeuchi, K.; Yokoo, J. (2001). "Paramyxovirus accessory proteins as interferon antagonists". Microbiology and Immunology. 45 (12): 787–800. doi:10.1111/j.1348-0421.2001.tb01315.x. ISSN 0385-5600. Archived from the original on 11 February 2022. Retrieved 26 March 2023.
- ↑ Morrison, T. G. (March 2001). "The three faces of paramyxovirus attachment proteins". Trends in Microbiology. 9 (3): 103–105. doi:10.1016/s0966-842x(01)01959-x. ISSN 0966-842X. Archived from the original on 29 March 2023. Retrieved 29 March 2023.
- ↑ Fearns, Rachel; Plemper, Richard K (2017-04-15). "Polymerases of paramyxoviruses and pneumoviruses". Virus Research. 234: 87–102. doi:10.1016/j.virusres.2017.01.008. ISSN 0168-1702. PMC 5476513. PMID 28104450.
- ↑ Harmon, Shawn B.; Megaw, A. George; Wertz, Gail W. (January 2001). "RNA Sequences Involved in Transcriptional Termination of Respiratory Syncytial Virus". Journal of Virology. 75 (1): 36–44. doi:10.1128/JVI.75.1.36-44.2001. ISSN 0022-538X. PMC 113895. PMID 11119571.
- ↑ Jacques, J. P.; Kolakofsky, D. (1991-05-01). "Pseudo-templated transcription in prokaryotic and eukaryotic organisms". Genes & Development. 5 (5): 707–713. doi:10.1101/gad.5.5.707. ISSN 0890-9369. PMID 2026325. S2CID 37461543. Archived from the original on 2022-12-26. Retrieved 2023-01-30.
- ↑ Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang LF (2012). "Cedar virus: a novel Henipavirus isolated from Australian bats". PLOS Pathogens. 8 (8): e1002836. doi:10.1371/journal.ppat.1002836. PMC 3410871. PMID 22879820.
- ↑ "Global Measles Mortality, 2000--2008". www.cdc.gov. Archived from the original on 2021-06-28. Retrieved 2023-01-30.
- ↑ "CDC – HPIVs – Overview of Human Parainfluenza Viruses". www.cdc.gov. Archived from the original on May 25, 2016. Retrieved September 19, 2014.
- 1 2 Caserta, MT, ed. (September 2013). "Measles". Merck Manual Professional. Merck Sharp & Dohme Corp. Archived from the original on 23 March 2014. Retrieved 23 March 2014.
- ↑ "Measles (Red Measles, Rubeola)". Dept of Health, Saskatchewan. Archived from the original on 10 February 2015. Retrieved 10 February 2015.
- ↑ "Measles Fact sheet N°286". World Health Organization (WHO). November 2014. Archived from the original on 3 February 2015. Retrieved 4 February 2015.
- ↑ Conn's Current Therapy 2015. Elsevier Health Sciences. 2014. p. 153. ISBN 9780323319560. Archived from the original on 2017-09-08.
- 1 2 3 "Measles (Rubeola) Signs and Symptoms". Centers for Disease Control and Prevention (CDC). 3 November 2014. Archived from the original on 2 February 2015. Retrieved 5 February 2015.
- ↑ "Pinkbook: Mumps | CDC". www.cdc.gov. 21 September 2022. Archived from the original on 6 July 2016. Retrieved 21 March 2023.
- 1 2 Bailey's head and neck surgery—otolaryngology. Johnson, Jonas T., Rosen, Clark A., Bailey, Byron J., 1934- (5th ed.). Philadelphia: Wolters Kluwer Health /Lippincott Williams & Wilkins. 2013. ISBN 9781609136024. OCLC 863599053.
{{cite book}}: CS1 maint: others (link) - ↑ Hviid A, Rubin S, Mühlemann K (March 2008). "Mumps". The Lancet. 371 (9616): 932–44. doi:10.1016/S0140-6736(08)60419-5. PMID 18342688. Archived from the original on 13 September 2020. Retrieved 26 June 2020.
- ↑ Henrickson, KJ; Kuhn, SM; Savatski, LL (May 1994). "Epidemiology and cost of infection with human parainfluenza virus types 1 and 2 in young children". Clinical Infectious Diseases. 18 (5): 770–9. doi:10.1093/clinids/18.5.770. PMID 8075269.
- ↑ Denny, FW; Clyde WA, Jr (May 1986). "Acute lower respiratory tract infections in nonhospitalized children". The Journal of Pediatrics. 108 (5 Pt 1): 635–46. doi:10.1016/S0022-3476(86)81034-4. PMID 3009769.
- ↑ "Acute Respiratory Infections". WHO. Archived from the original on March 24, 2006. Retrieved 21 March 2012.
- ↑ Rendon-Marin, Santiago; da Fontoura Budaszewski, Renata; Canal, Cláudio Wageck; Ruiz-Saenz, Julian (7 March 2019). "Tropism and molecular pathogenesis of canine distemper virus". Virology Journal. 16 (1): 30. doi:10.1186/s12985-019-1136-6. ISSN 1743-422X. Archived from the original on 14 January 2023. Retrieved 23 March 2023.
- ↑ Di Guardo, G.; Marruchella, G.; Agrimi, U.; Kennedy, S. (March 2005). "Morbillivirus infections in aquatic mammals: a brief overview". Journal of Veterinary Medicine. A, Physiology, Pathology, Clinical Medicine. 52 (2): 88–93. doi:10.1111/j.1439-0442.2005.00693.x. ISSN 0931-184X. Archived from the original on 26 October 2022. Retrieved 24 March 2023.
- ↑ Ganar, Ketan; Das, Moushumee; Sinha, Sugandha; Kumar, Sachin (12 May 2014). "Newcastle disease virus: current status and our understanding". Virus Research. 184: 71–81. doi:10.1016/j.virusres.2014.02.016. ISSN 1872-7492. Archived from the original on 15 June 2022. Retrieved 24 March 2023.
- ↑ Barrett, T.; Rossiter, P. B. (1999). "Rinderpest: the disease and its impact on humans and animals". Advances in Virus Research. 53: 89–110. doi:10.1016/s0065-3527(08)60344-9. ISSN 0065-3527. Archived from the original on 26 December 2022. Retrieved 24 March 2023.
- ↑ "Henipaviruses - Chapter 4 - 2020 Yellow Book | Travelers' Health | CDC". wwwnc.cdc.gov. Archived from the original on 14 March 2023. Retrieved 25 March 2023.
- ↑ Sawatsky (2008). "Hendra and Nipah Virus". Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6. Archived from the original on 2016-08-20. Retrieved 2023-01-30.
- ↑ Samal, S.K. (2008). "Paramyxoviruses of Animals". Encyclopedia of Virology: 40–47. doi:10.1016/B978-012374410-4.00460-X. Archived from the original on 4 April 2023. Retrieved 4 April 2023.
- ↑ McCarthy AJ, Goodman SJ (January 2010). "Reassessing conflicting evolutionary histories of the Paramyxoviridae and the origins of respiroviruses with Bayesian multigene phylogenies". Infect. Genet. Evol. 10 (1): 97–107. doi:10.1016/j.meegid.2009.11.002. PMID 19900582.
- ↑ Larsen, Brendan B.; Gryseels, Sophie; Otto, Hans W.; Worobey, Michael (9 February 2022). "Evolution and Diversity of Bat and Rodent Paramyxoviruses from North America". Journal of Virology. 96 (3): e0109821. doi:10.1128/JVI.01098-21. ISSN 1098-5514. Archived from the original on 11 November 2022. Retrieved 27 March 2023.
- ↑ Pomeroy, Laura W.; Bjørnstad, Ottar N.; Holmes, Edward C. (2008). "The Evolutionary and Epidemiological Dynamics of the Paramyxoviridae". Journal of Molecular Evolution. 66 (2): 98–106. doi:10.1007/s00239-007-9040-x. ISSN 0022-2844. Archived from the original on 8 January 2023. Retrieved 31 March 2023.
External links
- ICTV Report: Paramyxoviridae Archived 2023-03-13 at the Wayback Machine
- Paramyxoviruses (1998) — morphology, genome, replication, pathogenesis (special access required)
- "Hendra virus has a growing family tree". CSIRO Paramyxovirus press release. 2001. Archived from the original on 2007-08-04.
- Animal viruses Archived 2010-03-28 at the Wayback Machine
- Paramyxoviridae Genomes Viral Bioinformatics Resource Center
- Viralzone: Paramyxoviridae Archived 2017-04-29 at the Wayback Machine
- Virus Pathogen Database and Analysis Resource (ViPR): Paramyxoviridae Archived 2022-06-03 at the Wayback Machine