Antigenic shift
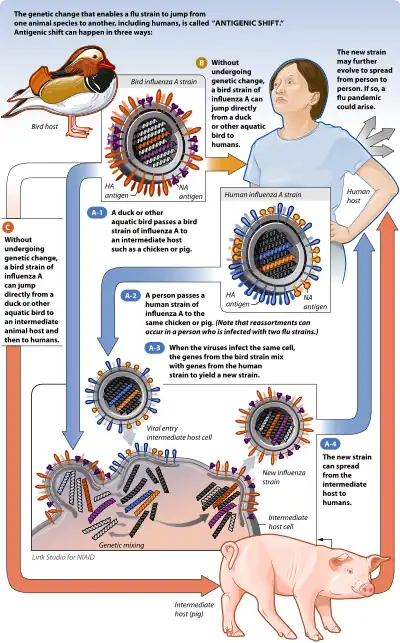
Antigenic shift is the process by which two or more different strains of a virus, or strains of two or more different viruses, combine to form a new subtype having a mixture of the surface antigens of the two or more original strains. The term is often applied specifically to influenza, as that is the best-known example, but the process is also known to occur with other viruses, such as visna virus in sheep.[1]
Antigenic shift is contrasted with antigenic drift, which is the natural mutation over time of known strains of influenza (or other things, in a more general sense) which may lead to a loss of immunity, or in vaccine mismatch. Antigenic drift occurs in all types of influenza including influenza A, influenza B and influenza C. Antigenic shift, however, occurs only in influenza A because it infects more than just humans.[2] Affected species include other mammals and birds, giving influenza A the opportunity for a major reorganization of surface antigens. Influenza B and C principally infect humans, minimizing the chance that a reassortment will change its phenotype drastically.[3]
In 1940s, Maurice Hilleman discovered antigenic shift, which is important for the emergence of new viral pathogens as it is a pathway that viruses may follow to enter a new niche.[4][5] It could occur with primate viruses and may be a factor for the appearance of new viruses in the human species such as HIV. HIV itself does not undergo reassortment/antigenic shift due to the structure of its genome, but it does recombine freely and via superinfection, so HIV can produce recombinant HIV strains that differ significantly from their ancestors.[6][7]
Transmission of influenza viruses from non-human animals to people
Influenza A viruses are found in many different animals, including ducks, chickens, pigs, humans, whales, horses, and seals.[3] Influenza B viruses circulate widely principally among humans, though it has recently been found in seals.[8] Flu strains are named after their types of hemagglutinin and neuraminidase surface proteins (of which there are 18 and 9 respectively), so they will be called, for example, H3N2 for type-3 hemagglutinin and type-2 neuraminidase. Some strains of avian influenza (from which all other strains of influenza A are believed to stem[2]) can infect pigs or other mammalian hosts. When two different strains of influenza infect the same cell simultaneously, their protein capsids and lipid envelopes are removed, exposing their RNA, which is then transcribed to mRNA. The host cell then forms new viruses that combine their antigens; for example, H3N2 and H5N1 can form H5N2 this way. Because the human immune system has difficulty recognizing the new influenza strain, it may be highly dangerous, and result in a new pandemic.[3]
Influenza viruses which have undergone antigenic shift have caused the Asian Flu pandemic of 1957, the Hong Kong Flu pandemic of 1968, and the Swine Flu scare of 1976. Until recently, such combinations were believed to have caused the infamous Spanish flu outbreak of 1918 which killed 40~100 million people worldwide. However, more recent research suggests the 1918 pandemic was caused by the antigenic drift of a fully avian virus to a form that could infect humans efficiently.[9][10] The most recent 2009 H1N1 outbreak was a result of antigenic shift and reassortment between human, avian, and swine viruses.[11]
Role of animals in Influenza antigenic shift
Pigs are especially important in antigenic shift of influenza viruses. Because pigs can be infected with strains of influenza that infect various other species of animals, they act as 'mixing pots' for the virus. When multiple virus strains, such as a duck and human influenza strain, infect the same pig, antigenic shift is likely to occur. While most of the virus strains resulting from this will be dead-end strains, a few have the potential to become pandemic viruses.[12]
Antigenic shift versus drift
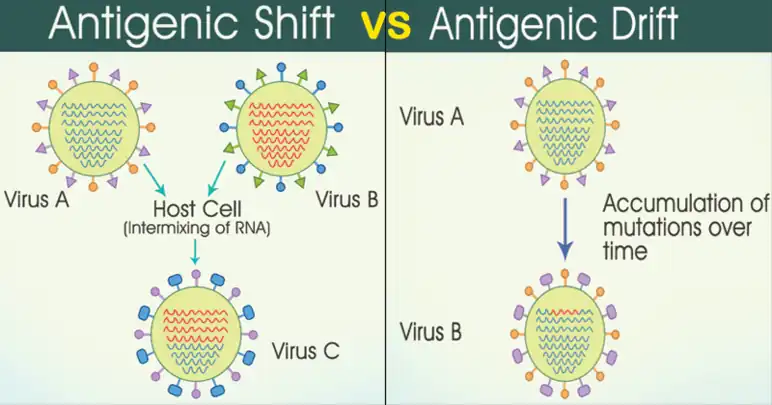
The difference between antigenic shift and antigenic drift are as follows:
- Antigenic shift:This happens when unique surface antigens from a strain that didn't exist before in the human population appears in a presently circulating strain via recombination[13]
- Antigenic drift: This alludes to the gradual buildup of point mutations during the annual spread of influenza as a result of error rates that have to do with RNA-dependent RNA polymerase in the course of viral replication.[14]
Influenza (flu virus) has the natural ability to change via antigenic shift or by antigenic drift[15]
Antigenic drift has been observed in SARS-CoV-2 (COVID-19 pandemic), according to a recent 2021 article[16]
See also
Notes
- ↑ Narayan, O; Griffin, DE; Chase, J (1977). "Antigenic shift of visna virus in persistently infected sheep". Science. 197 (4301): 376–378. Bibcode:1977Sci...197..376N. doi:10.1126/science.195339. PMID 195339.)
- 1 2 Treanor, John (15 January 2004). "Influenza vaccine--outmaneuvering antigenic shift and drift". New England Journal of Medicine. 350 (3): 218–220. doi:10.1056/NEJMp038238. PMID 14724300.
- 1 2 3 Zambon, Maria C. (November 1999). "Epidemiology and pathogenesis of influenza". Journal of Antimicrobial Chemotherapy. 44 (Supp B): 3–9. doi:10.1093/jac/44.suppl_2.3. PMID 10877456.
- ↑ Oransky, Ivan (14 May 2005). "Maurice R Hilleman". The Lancet. 365 (9472): 1682. doi:10.1016/S0140-6736(05)66536-1. ISSN 0140-6736. PMID 15912596. S2CID 46630955.
- ↑ Kurth, Reinhard (April 2005). "Maurice R. Hilleman (1919–2005)". Nature. 434 (7037): 1083. doi:10.1038/4341083a. ISSN 1476-4687. PMID 15858560. S2CID 26364385.
- ↑ Fanales-Belasio, Emanuele; Raimondo, Mariangela; Suligoi, Barbara; Buttò, Stefano (2010). "HIV virology and pathogenetic mechanisms of infection: a brief overview". Annali dell'Istituto Superiore Di Sanita. 46 (1): 5–14. doi:10.4415/ANN_10_01_02. ISSN 2384-8553. Archived from the original on 7 June 2021. Retrieved 9 September 2021.
- ↑ "Human Immunodeficiency Virus (HIV) Infection - Infections". Merck Manuals Consumer Version. Archived from the original on 4 June 2021. Retrieved 10 September 2021.
- ↑ Carrington, Damian (11 May 2000). "Seals pose influenza threat". BBC. Archived from the original on 28 July 2003. Retrieved 31 August 2021.
- ↑ Aoki, FY; Sitar, DS (January 1988). "Clinical pharmacokinetics of amantadine hydrochloride". Clinical Pharmacokinetics. 14 (1): 35–51. doi:10.2165/00003088-198814010-00003. PMID 3280212. S2CID 38462095.
- ↑ Johnson, NP; Mueller, J (Spring 2002). "Updating the accounts: global mortality of the 1918-1920 "Spanish" influenza pandemic". Bulletin of the History of Medicine. 76 (1): 105–115. doi:10.1353/bhm.2002.0022. PMID 11875246. S2CID 22974230.
- ↑ Smith, G. J. D.; Vijaykrishna, D.; Bahl, J.; Lycett, S. J.; Worobey, M.; Pybus, O. G.; Ma, S. K.; Cheung, C. L.; Raghwani, J.; Bhatt, S.; Peiris, J. S. M.; Guan, Y.; Rambaut, A. (2009). "Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic". Nature. 459 (7250): 1122–1125. Bibcode:2009Natur.459.1122S. doi:10.1038/nature08182. PMID 19516283.
- ↑ "Key Facts about Human Infections with Variant Viruses | CDC". www.cdc.gov. 3 January 2019. Archived from the original on 16 July 2021. Retrieved 15 November 2020.
- ↑ Greenbaum, Benjamin D; Ghedin, Elodie (August 2015). "Viral evolution: beyond drift and shift". Current Opinion in Microbiology. 26: 109–115. doi:10.1016/j.mib.2015.06.015. ISSN 1369-5274. Archived from the original on 8 September 2021. Retrieved 7 September 2021.
- ↑ Infectious diseases (3rd ed.). [Edinburgh]: Mosby/Elsevier. 2010. pp. 3–29. ISBN 978-0-323-04579-7. Archived from the original on 19 April 2021. Retrieved 8 September 2021.
- ↑ "How Flu Viruses Can Change". Centers for Disease Control and Prevention. 15 October 2019. Archived from the original on 18 March 2021. Retrieved 11 September 2021.
- ↑ Yuan, Meng; Huang, Deli; Lee, Chang-Chun D.; Wu, Nicholas C.; Jackson, Abigail M.; Zhu, Xueyong; Liu, Hejun; Peng, Linghang; van Gils, Marit J.; Sanders, Rogier W.; Burton, Dennis R.; Reincke, S. Momsen; Prüss, Harald; Kreye, Jakob; Nemazee, David; Ward, Andrew B.; Wilson, Ian A. (20 May 2021). "Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants". Science (New York, N.y.). doi:10.1126/science.abh1139. ISSN 0036-8075. Archived from the original on 28 August 2021. Retrieved 11 September 2021.
Further reading
- Bouvier, Nicole M.; Palese, Peter (September 2008). "The biology of influenza viruses". Vaccine. 26 Suppl 4 (Suppl 4): D49–53. doi:10.1016/j.vaccine.2008.07.039. PMC 3074182. PMID 19230160.
External links
- Superflu: Antigenic shift in Influenza Archived 15 September 2016 at the Wayback Machine Archived 15 September 2016 at the Wayback Machine
