Cancer pharmacogenomics
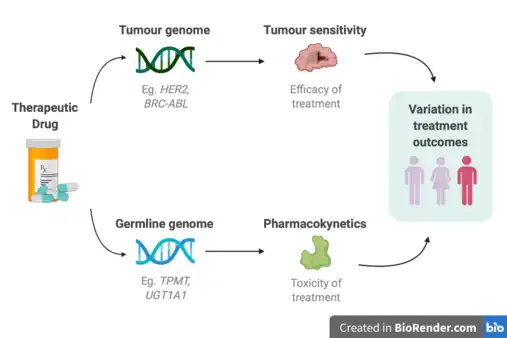
Cancer pharmacogenomics is the study of how variances in the genome influences an individual’s response to different cancer drug treatments. It is a subset of the broader field of pharmacogenomics, which is the area of study aimed at understanding how genetic variants influence drug efficacy and toxicity.[1]
Cancer is a genetic disease where changes to genes can cause cells to grow and divide out of control. Each cancer can have a unique combination of genetic mutations, and even cells within the same tumour may have different genetic changes. In clinical settings, it has commonly been observed that the same types and doses of treatment can result in substantial differences in efficacy and toxicity across patients.[2][3] Thus, the application of pharmacogenomics within the field of cancer can offer key advantages for personalizing cancer therapy, minimizing treatment toxicity, and maximizing treatment efficacy. This can include choosing drugs that target specific mutations within cancer cells, identifying patients at risk for severe toxicity to a drug, and identifying treatments that a patient is most likely to benefit from.[4] Applying pharmacogenomics within cancer has considerable differences compared to other complex diseases, as there are two genomes that need to be considered - the germline and the tumour. The germline genome considers inter-individual inherited genetic variations, and the tumour genome considers any somatic mutations that accrue as a cancer evolves.[5] The accumulation of somatic mutations within the tumour genome represents variation in disease, and plays a major role in understanding how individuals will respond to treatments. Additionally, the germline genome affects toxicity reactions to a specific treatment due to its influence on drug exposure. Specifically, pharmacokinetic genes participate in the inactivation and elimination of active compounds.[6] Therefore, differences within the germline genome should also be considered.[5][7][8]
Strategies
Advances in cancer diagnostics and treatment have shifted the use of traditional methods of physical examination, in vivo, and histopathological analysis to assessment of cancer drivers, mutations, and targetable genomic biomarkers.[9] There are an increasing number of genomic variants being studied and identified as potential therapeutically actionable targets and drug metabolism modifiers.[10][11] Thus, a patient's genomic information, in addition to information about the patient's tumour, can be used to determine a personalized approach to cancer treatment.[9][12]
Cancer-driven DNA alterations
Cancer-driven DNA alterations can include somatic DNA mutations and inherited DNA variants. They are not a direct focus of pharmacogenomic studies, but they can have an impact on pharmacogenomic strategies.[9] These alterations can affect the pharmacokinetics and pharmacodynamics of metabolic pathways, making them potentially actionable drug-targets.
As whole-genome technologies continue to advance, there will be increased opportunities to discover mutations and variants that are involved in tumour progression, response to therapy, and drug-metabolism.
Polymorphism search
Candidate polymorphism search refers to finding polymorphic DNA sequences within specific genes that are candidates for certain traits. Within pharmacogenomics, this method tries to resolve pharmacokinetic or pharmacodynamic traits of a compound to a candidate polymorphism level.[9][13] This type of information can contribute to selecting effective therapeutic strategies for a patient.
To understand the potential functional impact of a polymorphic DNA sequence, gene silencing can be used. Previously, siRNAs have been commonly used to suppress gene expressions, but more recently, siRNA have been suggested for use in studying and developing therapeutics.[14][15]
Another new method being applied is Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR). CRISPR, combined with the Cas9 enzyme, form the basis for the technology known as CRISPR-Cas9. This system can recognize and cleave specific DNA sequences, and thus is a powerful system for gene silencing purposes.[16]
Pathway search
An extension on the previous strategies is candidate pathway search. This type of analysis considers a group of related genes, whose altered function may have an effect on therapy, rather than solely focusing on one gene. It can provide insight into additional information such as gene-gene interactions, epistatic effects, or influences from cis-regulatory elements.[9][17] These all contribute to understanding variations in drug efficacy and toxicity between patients.
Whole-Genome Strategies
Advancements in the cost and throughput of sequencing technologies is making it possible to perform whole-genome sequencing at higher rates. The ability to perform whole-genome analysis for cancer patients can aid in identifying markers of predisposition to drug toxicity and efficacy.[18] Strategies for pharmacogenomic discovery using whole-genome sequences include targeting frequently mutated gene stretches (known as hotspots) to identify markers of prognostic and diagnostic significance, or targeting specific genes that are known to be associated with a particular disease.[9]
Gene target examples
HER2
HER2 is an established therapeutic target within breast cancer, and the activation of HER2 is observed in approximately 20% of breast cancers as a result of overexpression.[19][20] Trastuzumab, the first HER2-targeted drug developed in 1990, interferes with HER2 signalling. In 2001, a study showed that adding trastuzumab to chemotherapy improved overall survival in women with HER2-positive metastatic breast cancer.[21] Then, in 2005, it was shown that trastuzumab is effective as an adjuvant treatment in women with early-stage breast cancer.[19][22] Thus, trastuzumab has been a standard-of-care treatment in both metastatic and early stage HER2-positive breast cancer cases. Many genome sequencing studies have also revealed that other cancer tumours had HER2 alterations, including overexpression, amplifications and other mutations.[23][24][25][26] Because of this, there has been a lot of interest in studying the efficacy of HER2-targeted therapies within a range of cancer types, including bladder, colorectal, and gastro-esophageal.
BRC-ABL
The majority of chronic myelogenous leukemia cases are caused by a rearrangement between chromosomes 9 and 22. This results in the fusion of the genes BCR and ABL. This atypical gene fusion encodes for unregulated tyrosine kinase activity, which results in the rapid and continuous division of white blood cells.[20][27] Drugs known as tyrosine kinase inhibitors target BCR-ABL, and are the standard treatment for chronic myelogenous leukemia. Imatinib was the first tyrosine kinase inhibitor discovered with high specificity for targeting BCR-ABL.[28] However, after imatinib was used as the first-line therapy, several BCR-ABL-dependent and BCR-ABL-independent mechanisms of resistance developed. Thus, new second-line and third-line drugs have also been developed to address new, mutated forms of BCR-ABL. These include dasatinib, nilotinib, bosutinib, and ponatinib.[27]
Pharmacokinetic genes
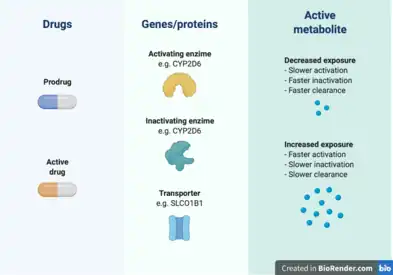
Cancer pharmacogenomics has also contributed to the understanding of how pharmacokinetic genes affect the exposure to cancer drugs, which can help predict patient sensitivity to treatment toxicity.[6] Some of these findings have been successfully translated into clinical practice in the form of professional guidelines from the Clinical Pharmacogenomics Implementation Consortium (CPIC) or other institutions.[29]
TPMT
The TPMT gene encodes for the thiopurine S-methyltransferase (TPMT) enzyme. It participates in the S-methylation of thiopurine drugs, which include 6-mercaptopurine, 6-thioguanine, and Azathioprine.[30] The first two drugs are indicated for leukemias and lymphomas, while Azathioprine is used in nonmalignant conditions such as Crohn’s disease. These purine antimetabolites are activated in the form of thioguanine nucleotides that affect DNA replication when incorporated into DNA.[6] This activation occurs through hypoxanthine phosphoribosyltransferase to 6-thioguanosines (6-TGN), and the resulting antimetabolites are inactivated by TPMT.[29] It has been established that the TPMT genotype of a patient can affect the levels of exposure to the active metabolites, which has an impact in the treatment toxicity and efficacy.[31][32] Specifically, TPMT-deficient patients, such as those homozygous for the *2 and *3 alleles, can experience myelosuppression up to pancytopenia.[33][29] In a study on 1214 European Caucasian individuals, a trimodal distribution of TPMT genotypes was found, with 89.5% normal-to-high methylators, 9.9% intermediates, and 0.6% deficient methylators[33] CPIC guidelines recommend a dose reduction of 5-10% of the standard dose and a lower frequency of application in individuals that are TPMT poor metabolizers.[34]
DPD
The dihydropyrimidine dehydrogenase (DPD) protein is responsible for the inactivation of more than 80% of the anticancer drug 5-Fluorouracil (5-FU) in the liver. This drug is commonly used in colorectal cancer treatment, and increased exposure to it can cause myelosuppression, mucositis, neurotoxicity, hand-foot syndrome, and diarrhea.[29] The genotype of DPYD (the gene that codes for DPD) has been linked to severe 5-FU toxicities in several studies summarized in meta-analyses.[35][36][37] The CPIC has provided guidelines for implementation of DPYD pharmacogenetics, indicating that homozygote carriers of low-activity variants should be prescribed an alternative drug, while heterozygotes should receive half of the normal dose.[38]
UGT1A1
The UDP glucuronosyltransferase 1A1 (UGT1A1) is an hepatic enzyme involved in the glucoronidation of exogenous and endogenous substrates, such as bilirubin.[6][39] There have been over 100 variants identified in UGT1A1 and some mutations are implicated Gilbert syndrome and Cringler-Najjar syndrome. Two variants in particular, UGT1A1*28 and UGT1A1*6, are associated with the pharmacogenomics of irinotecan chemotherapy. A UGT1A1*28 allele means the presence of 7 TA repeats in the promoter sequence of the gene, instead of the normal 6 repeats.[6] The allele UGT1A1*6 is characterized by a SNP in exon 1.[40]
Irinotecan is a prodrug[6] used in the treatment of many solid tumours, including colorectal, pancreatic, and lung cancer.[41] Irinotecan is metabolized into its active compound SN-38, which inhibits the enzyme topoisomerase-1, involved in DNA replication.[42] This active metabolite is inactivated after glucoronidation, mainly performed by UGT1A1.[39] High exposure to SN-38 can result in neutropenia and gastrointestinal toxicity.[6] The decreased activity of UGT1A1 in UGT1A1*28 individuals has been found to increase exposure to the active compound and toxicity.[43][44] For UGT1A1*6, this relationship is more controversial, with some studies finding it can predict irinotecan toxicity while others don’t.[40] Previous prospective studies for assessing the adequate dose of irinotecan in Asians have supported the usage of lower doses in patients with both of UGT1A1*28 and UGT1A1*6.[45][46] The results from these and other pharmacogenomics studies have been translated into clinical guidelines from organizations in USA, Canada, France, The Netherlands, and Europe.[41] All of these institutions recommend a dose reduction in UGT1A1*28 patients.
Challenges
One of the biggest challenges in using pharmacogenomics to study cancer is the difficulty in conducting studies in humans. Drugs used for chemotherapy are too toxic to give to healthy individuals, which makes it difficult to perform genetic studies between related individuals.[5] Furthermore, some mutations occur at high frequencies, whereas others occur at very low frequencies, so there is often a need to screen a large number of patients in order to identify those with a particular genetic marker. And, although genomic-driven analyses is effective for stratifying patients and identifying possible treatment options, it is often difficult for laboratories to get reimbursed for these genomic sequencing tests. Thus, tracking clinical outcomes for patients whom undergo sequencing is key to demonstrating both the clinical utility and cost-effectiveness of pharmacogenomics within cancer.[47]
Another challenge is that cancer patients are often treated with different combinations and dosages of drugs, so finding a large sample of patients that have been treated the same way is rare. So, studying the pharmacogenomics of a specific drug of interest is difficult, and, because additional identical trials may not be feasible, it can be difficult to replicate discoveries.[1]
Furthermore, studies have shown that drug efficacy and toxicity are likely multigenic traits. Since pathways contain multiple genes, various combinations of driver mutations could promote tumour progression.[47][48][49] This can make it difficult to distinguish between functional driver mutations versus random, nonfunctional mutations.[50]
Future
With new tools and technologies continuing to develop, there are growing opportunities to analyze cancer at the single-cell level. Corresponding approaches with whole-genome sequencing can also be applied to single-cell sequences and analyses. This level of pharmacogenomics has implications in personalized medicine, as single-cell RNA sequencing and genotyping can characterize subclones of the same tumour,[9] and lead to the identification therapy-resistant cells, as well as their corresponding pathways.[51]
As the ability to analyze and profile cancers continues to improve, so will the therapies developed to treat them. And, with increasing attention being given to whole-genome sequencing and single-cell sequencing, there will be a growing amount of pharmacogenomic data to analyze. These analyses will rely on new and improved bioinformatics tools to help identify targetable genes and pathways, to help select safer and more effect therapies for cancer patients.
References
- 1 2 Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ (January 2013). "Cancer pharmacogenomics: strategies and challenges". Nature Reviews. Genetics. 14 (1): 23–34. doi:10.1038/nrg3352. PMC 3668552. PMID 23183705.
- ↑ Evans WE, Relling MV (October 1999). "Pharmacogenomics: translating functional genomics into rational therapeutics". Science. 286 (5439): 487–91. doi:10.1126/science.286.5439.487. PMID 10521338.
- ↑ Fagerlund TH, Braaten O (February 2001). "No pain relief from codeine...? An introduction to pharmacogenomics". Acta Anaesthesiologica Scandinavica. 45 (2): 140–9. PMID 11167158.
- ↑ "What Is Cancer?". National Cancer Institute. 2007-09-17. Retrieved 2020-02-26.
- 1 2 3 Moen EL, Godley LA, Zhang W, Dolan ME (2012). "Pharmacogenomics of chemotherapeutic susceptibility and toxicity". Genome Medicine. 4 (11): 90. doi:10.1186/gm391. PMC 3580423. PMID 23199206.
- 1 2 3 4 5 6 7 Hertz DL, Rae J (2015-01-14). "Pharmacogenetics of cancer drugs". Annual Review of Medicine. 66 (1): 65–81. doi:10.1146/annurev-med-053013-053944. PMID 25386932.
- ↑ Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Badner JA (June 2004). "Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity". Cancer Research. 64 (12): 4353–6. doi:10.1158/0008-5472.CAN-04-0340. PMID 15205351.
- ↑ Wen Y, Gorsic LK, Wheeler HE, Ziliak DM, Huang RS, Dolan ME (August 2011). "Chemotherapeutic-induced apoptosis: a phenotype for pharmacogenomics studies". Pharmacogenetics and Genomics. 21 (8): 476–88. doi:10.1097/FPC.0b013e3283481967. PMC 3134538. PMID 21642893.
- 1 2 3 4 5 6 7 Concetta Crisafulli, Concetta; Romeo, Petronilla Daniela; Calabrò, Marco; Epasto, Ludovica Martina; Alberti, Saverio (2019). "Pharmacogenetic and pharmacogenomic discovery strategies". Cancer Drug Resistance. 2 (2): 225–241. doi:10.20517/cdr.2018.008. Retrieved 2020-02-26.
- ↑ Cascorbi I, Bruhn O, Werk AN (May 2013). "Challenges in pharmacogenetics". European Journal of Clinical Pharmacology. 69 Suppl 1: 17–23. doi:10.1007/s00228-013-1492-x. PMID 23640184. S2CID 2667130.
- ↑ El-Deiry WS, Goldberg RM, Lenz HJ, Shields AF, Gibney GT, Tan AR, et al. (July 2019). "The current state of molecular testing in the treatment of patients with solid tumors, 2019". CA: A Cancer Journal for Clinicians. 69 (4): 305–343. doi:10.3322/caac.21560. PMC 6767457. PMID 31116423.
- ↑ Adams DR, Eng CM (October 2018). "Next-Generation Sequencing to Diagnose Suspected Genetic Disorders". The New England Journal of Medicine. 379 (14): 1353–1362. doi:10.1056/NEJMra1711801. PMID 30281996.
- ↑ Cockram J, White J, Zuluaga DL, Smith D, Comadran J, Macaulay M, et al. (December 2010). "Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome". Proceedings of the National Academy of Sciences of the United States of America. 107 (50): 21611–6. Bibcode:2010PNAS..10721611C. doi:10.1073/pnas.1010179107. PMC 3003063. PMID 21115826.
- ↑ Lambeth LS, Smith CA (2013). "Short hairpin RNA-mediated gene silencing". SiRNA Design. Methods in Molecular Biology. Vol. 942. pp. 205–32. doi:10.1007/978-1-62703-119-6_12. ISBN 978-1-62703-118-9. PMID 23027054.
- ↑ Moore CB, Guthrie EH, Huang MT, Taxman DJ (2010). "Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown". RNA Therapeutics. Methods in Molecular Biology. Vol. 629. pp. 141–58. doi:10.1007/978-1-60761-657-3_10. ISBN 978-1-60761-656-6. PMC 3679364. PMID 20387148.
- ↑ Zhang F, Wen Y, Guo X (September 2014). "CRISPR/Cas9 for genome editing: progress, implications and challenges". Human Molecular Genetics. 23 (R1): R40-6. doi:10.1093/hmg/ddu125. PMID 24651067.
- ↑ Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, et al. (January 2019). "Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks". Cell. 176 (1–2): 361–376.e17. doi:10.1016/j.cell.2018.11.022. PMC 6329648. PMID 30580963.
- ↑ Rabbani B, Nakaoka H, Akhondzadeh S, Tekin M, Mahdieh N (May 2016). "Next generation sequencing: implications in personalized medicine and pharmacogenomics". Molecular BioSystems. 12 (6): 1818–30. doi:10.1039/C6MB00115G. PMID 27066891.
- 1 2 Oh DY, Bang YJ (January 2020). "HER2-targeted therapies - a role beyond breast cancer". Nature Reviews. Clinical Oncology. 17 (1): 33–48. doi:10.1038/s41571-019-0268-3. PMID 31548601. S2CID 202729132.
- 1 2 "Pharmacogenomics and cancer". yourgenome. Retrieved 2020-02-26.
- ↑ Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. (March 2001). "Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2". The New England Journal of Medicine. 344 (11): 783–92. doi:10.1056/NEJM200103153441101. PMID 11248153.
- ↑ Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. (October 2005). "Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer". The New England Journal of Medicine. 353 (16): 1659–72. doi:10.1056/NEJMoa052306. hdl:10722/251817. PMID 16236737.
- ↑ Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. (August 2010). "Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial". Lancet. 376 (9742): 687–97. doi:10.1016/S0140-6736(10)61121-X. PMID 20728210. S2CID 8825706.
- ↑ Yoshida H, Shimada K, Kosuge T, Hiraoka N (April 2016). "A significant subgroup of resectable gallbladder cancer patients has an HER2 positive status". Virchows Archiv. 468 (4): 431–9. doi:10.1007/s00428-015-1898-1. PMID 26758058. S2CID 23543906.
- ↑ Seo AN, Kwak Y, Kim DW, Kang SB, Choe G, Kim WH, Lee HS (2014). "HER2 status in colorectal cancer: its clinical significance and the relationship between HER2 gene amplification and expression". PLOS ONE. 9 (5): e98528. Bibcode:2014PLoSO...998528S. doi:10.1371/journal.pone.0098528. PMC 4039475. PMID 24879338.
- ↑ Yan M, Schwaederle M, Arguello D, Millis SZ, Gatalica Z, Kurzrock R (March 2015). "HER2 expression status in diverse cancers: review of results from 37,992 patients". Cancer and Metastasis Reviews. 34 (1): 157–64. doi:10.1007/s10555-015-9552-6. PMC 4368842. PMID 25712293.
- 1 2 Rossari F, Minutolo F, Orciuolo E (June 2018). "Past, present, and future of Bcr-Abl inhibitors: from chemical development to clinical efficacy". Journal of Hematology & Oncology. 11 (1): 84. doi:10.1186/s13045-018-0624-2. PMC 6011351. PMID 29925402.
- ↑ Eck MJ, Manley PW (April 2009). "The interplay of structural information and functional studies in kinase drug design: insights from BCR-Abl". Current Opinion in Cell Biology. 21 (2): 288–95. doi:10.1016/j.ceb.2009.01.014. PMID 19217274.
- 1 2 3 4 Cascorbi I, Werk AN (January 2017). "Advances and challenges in hereditary cancer pharmacogenetics". Expert Opinion on Drug Metabolism & Toxicology. 13 (1): 73–82. doi:10.1080/17425255.2017.1233965. PMID 27603572. S2CID 38766779.
- ↑ Weinshilboum RM (January 1992). "Methylation pharmacogenetics: thiopurine methyltransferase as a model system". Xenobiotica; the Fate of Foreign Compounds in Biological Systems. 22 (9–10): 1055–71. doi:10.3109/00498259209051860. PMID 1441597.
- ↑ Lennard L, Lilleyman JS, Van Loon J, Weinshilboum RM (July 1990). "Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia". Lancet. 336 (8709): 225–9. doi:10.1016/0140-6736(90)91745-V. PMID 1973780. S2CID 25426385.
- ↑ Black AJ, McLeod HL, Capell HA, Powrie RH, Matowe LK, Pritchard SC, et al. (November 1998). "Thiopurine methyltransferase genotype predicts therapy-limiting severe toxicity from azathioprine". Annals of Internal Medicine. 129 (9): 716–8. doi:10.7326/0003-4819-129-9-199811010-00007. PMID 9841604. S2CID 9267887.
- 1 2 Schaeffeler E, Fischer C, Brockmeier D, Wernet D, Moerike K, Eichelbaum M, et al. (July 2004). "Comprehensive analysis of thiopurine S-methyltransferase phenotype-genotype correlation in a large population of German-Caucasians and identification of novel TPMT variants". Pharmacogenetics. 14 (7): 407–17. doi:10.1097/01.fpc.0000114745.08559.db. PMID 15226673.
- ↑ Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui CH, Yee SW, et al. (March 2011). "Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing". Clinical Pharmacology and Therapeutics. 89 (3): 387–91. doi:10.1038/clpt.2010.320. PMC 3098761. PMID 21270794.
- ↑ Rosmarin D, Palles C, Church D, Domingo E, Jones A, Johnstone E, et al. (April 2014). "Genetic markers of toxicity from capecitabine and other fluorouracil-based regimens: investigation in the QUASAR2 study, systematic review, and meta-analysis". Journal of Clinical Oncology. 32 (10): 1031–9. doi:10.1200/JCO.2013.51.1857. PMC 4879695. PMID 24590654.
- ↑ Terrazzino S, Cargnin S, Del Re M, Danesi R, Canonico PL, Genazzani AA (August 2013). "DPYD IVS14+1G>A and 2846A>T genotyping for the prediction of severe fluoropyrimidine-related toxicity: a meta-analysis". Pharmacogenomics. 14 (11): 1255–72. doi:10.2217/pgs.13.116. PMID 23930673.
- ↑ Meulendijks D, Henricks LM, Sonke GS, Deenen MJ, Froehlich TK, Amstutz U, et al. (December 2015). "Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: a systematic review and meta-analysis of individual patient data". The Lancet. Oncology. 16 (16): 1639–50. doi:10.1016/S1470-2045(15)00286-7. PMID 26603945.
- ↑ Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, Diasio RB, Schwab M (December 2013). "Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing". Clinical Pharmacology and Therapeutics. 94 (6): 640–5. doi:10.1038/clpt.2013.172. PMC 3831181. PMID 23988873.
- 1 2 Takano M, Sugiyama T (2017-02-28). "UGT1A1 polymorphisms in cancer: impact on irinotecan treatment". Pharmacogenomics and Personalized Medicine. 10: 61–68. doi:10.2147/pgpm.s108656. PMC 5338934. PMID 28280378.
- 1 2 Zhang X, Yin JF, Zhang J, Kong SJ, Zhang HY, Chen XM (July 2017). "UGT1A1*6 polymorphisms are correlated with irinotecan-induced neutropenia: a systematic review and meta-analysis". Cancer Chemotherapy and Pharmacology. 80 (1): 135–149. doi:10.1007/s00280-017-3344-3. PMID 28585035. S2CID 8697984.
- 1 2 de Man FM, Goey AK, van Schaik RH, Mathijssen RH, Bins S (October 2018). "Individualization of Irinotecan Treatment: A Review of Pharmacokinetics, Pharmacodynamics, and Pharmacogenetics". Clinical Pharmacokinetics. 57 (10): 1229–1254. doi:10.1007/s40262-018-0644-7. PMC 6132501. PMID 29520731.
- ↑ Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y (March 1999). "Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes". The EMBO Journal. 18 (5): 1397–406. doi:10.1093/emboj/18.5.1397. PMC 1171229. PMID 10064605.
- ↑ Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D'Andrea M, et al. (July 2006). "The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer". Journal of Clinical Oncology. 24 (19): 3061–8. doi:10.1200/JCO.2005.05.5400. PMID 16809730.
- ↑ Marcuello E, Altés A, Menoyo A, Del Rio E, Gómez-Pardo M, Baiget M (August 2004). "UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer". British Journal of Cancer. 91 (4): 678–82. doi:10.1038/sj.bjc.6602042. PMC 2364770. PMID 15280927.
- ↑ Hazama S, Nagashima A, Kondo H, Yoshida S, Shimizu R, Araki A, et al. (March 2010). "Phase I study of irinotecan and doxifluridine for metastatic colorectal cancer focusing on the UGT1A1*28 polymorphism". Cancer Science. 101 (3): 722–7. doi:10.1111/j.1349-7006.2009.01428.x. PMID 20028383.
- ↑ "IN THIS ISSUE". Japanese Journal of Clinical Oncology. 41 (4): NP. 2011-04-01. doi:10.1093/jjco/hyr047. ISSN 0368-2811.
- 1 2 Patel JN (2016-07-12). "Cancer pharmacogenomics, challenges in implementation, and patient-focused perspectives". Pharmacogenomics and Personalized Medicine. 9: 65–77. doi:10.2147/pgpm.s62918. PMC 4948716. PMID 27471406.
- ↑ Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. (October 2010). "The patterns and dynamics of genomic instability in metastatic pancreatic cancer". Nature. 467 (7319): 1109–13. Bibcode:2010Natur.467.1109C. doi:10.1038/nature09460. PMC 3137369. PMID 20981101.
- ↑ Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. (March 2012). "Intratumor heterogeneity and branched evolution revealed by multiregion sequencing". The New England Journal of Medicine. 366 (10): 883–892. doi:10.1056/NEJMoa1113205. PMC 4878653. PMID 22397650.
- ↑ Stratton MR, Campbell PJ, Futreal PA (April 2009). "The cancer genome". Nature. 458 (7239): 719–24. Bibcode:2009Natur.458..719S. doi:10.1038/nature07943. PMC 2821689. PMID 19360079.
- ↑ Irish JM, Kotecha N, Nolan GP (February 2006). "Mapping normal and cancer cell signalling networks: towards single-cell proteomics". Nature Reviews. Cancer. 6 (2): 146–55. doi:10.1038/nrc1804. PMID 16491074. S2CID 13130810.