Hemagglutinin
In molecular biology, hemagglutinin (or haemagglutinin in British English) (from the Greek haima, 'blood' + Latin gluten, 'glue') is a glycoprotein which causes red blood cells (RBCs) to agglutinate or clump together. This is one of three steps in the more complex process of coagulation.
Agglutination mostly happens when adding influenza virus to red blood cells, as virologist George K. Hirst discovered in 1941. It can also occur with measles virus, parainfluenza virus and mumps virus, among others. Alfred Gottschalk proved in 1957 that hemagglutinin binds a virus to a host cell by attaching to sialic acids on carbohydrate side chains of cell-membrane glycoproteins and glycolipids.[1]
There are different types of hemagglutinin but, in general, two groups can be described, depending on how they act in different temperatures:
- Cold hemagglutinin: which can act in an optimal manner at temperatures reaching 4°C.
- Warm hemagglutinin: which can act in an optimal manner at temperatures reaching 37°C.
Antibodies[2] and lectins[3] are commonly known hemagglutinins.
Types
Examples include:
- Influenza hemagglutinin or haemagglutinin: a homotrimeric glycoprotein that is found on the surface of influenza viruses; it provides part of their infectivity.
- Measles hemagglutinin: a hemagglutinin produced by measles virus which encodes six structural proteins, of which two, hemagglutinin and fusion, are surface glycoproteins involved in attachment and entry.[4]
- Parainfluenza hemagglutinin-neuraminidase: a type of hemagglutinin-neuraminidase produced by parainfluenza which is closely associated with both human and veterinary disease.
- Mumps hemagglutinin-neuraminidase: a kind of hemagglutinin that the mumps virus (MuV) produces, which is the virus that causes mumps.
- The PH-E form of phytohaemagglutinin
Structure
The hemagglutinin in itself has a cylinder shape and it’s a quite small protein (13 nanometers long). It’s a glycoprotein formed by three identical subunits called monomers, so we can tell that it’s a homotrimer. These monomers are linked by two disulfide polypeptides: membrane-distal HA1 and the membrane-proximal HA2 which is much smaller. With the help of different studies, such as X-ray crystallography, and also different kinds of spectroscopic techniques, some researchers found that in the hemagglutinin membrane there is a majority of α-helical structures rather than β-pleated sheet. The trimer structure is linked to the membrane by little elastic chains which can reach to the detergent micelle in order to create a bundle of three α-helices that makes the structure more stable. These α-helices can adopt different angles in the structure, these ones can go from 0° up to 25° between each other. These different orientations are steady thanks to other types of bonds, such as hydrogen bonds, that also contribute to the stability of the glycoprotein.[5]
Uses in serology
- HIA (Hemagglutination Inhibition Assay):[6] is a serologic assay which can be used either to screen for antibodies using RBCs with known surface antigens or to identify RBCs surface antigens such as viruses or bacteria using a panel of known antibodies. This method, performed first by George K. Hirst in 1942, consists of mixing virus samples with serum dilutions so that antibodies have already binded the virus by the time RBCs are added to the mix. Consequently, those viruses bounded to antibodies will be unable to link RBCs, meaning that a test’s positive result due to hemagglutination has been inhibited. On the contrary, if hemagglutination occurs, the test will result negative.
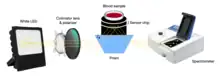
- Hemagglutination blood typing detection:[7] this method for on-site use consists of measuring both blood’s reflectance spectrum alone (non-agglutination) and that of blood mixed with antibody reagents (agglutination) using a waveguide-mode sensor. As a result, some differences in reflectance between the samples are observed and, once antibodies are added, blood types and Rh(D) typing are also possible to distinguish thanks to the waveguide-mode sensor. Besides, what is the most important about this technique is the fact that it is able to detect weak agglutinations, which are almost impossible to detect for human eyes.
- Using anti-A and anti-B antibodies that bind specifically to either the A or to the B blood group surface antigens on RBCs it is possible to test a small sample of blood and determine the ABO blood group (or blood type) of an individual. The negative points are that it does not identify the Rh(D) typing as well as the instruments needed lack the required usability or portability.
- The bedside card method of blood grouping relies on visual agglutination to determine an individual's blood group. The card has dried blood group antibody reagents fixed onto its surface and a drop of the individual's blood is placed on each area on the card. The presence or absence of visual agglutination enables a quick and convenient method of determining the ABO and Rhesus status of the individual. As this technique depends on human eyes, it is less reliable than the blood typing based on waveguide-mode sensors.
- Agglutination of red blood cells is used in the Coombs test.
See also
- Cold agglutinin disease
- Hemagglutination assay
- Neuraminidase
- Influenza hemagglutinin (HA)
- Agglutination
References
- ↑ Henry, Ronnie; Murphy, Frederick A. (October 2018). "Etymologia: Hemagglutinin and Neuraminidase". Emerging Infectious Diseases. 24 (10): 1849. doi:10.3201/eid2410.ET2410. PMC 6154157.
- ↑ "hemagglutinin" at Dorland's Medical Dictionary
- ↑ Hemagglutinins at the US National Library of Medicine Medical Subject Headings (MeSH)
- ↑ Pan CH, Jimenez GS, Nair N (August 21, 2014) [August, 2008]. "Use of Vaxfectin Adjuvant with DNA Vaccine Encoding the Measles Virus Hemagglutinin and Fusion Proteins Protects Juvenile and Infant Rhesus Macaques against Measles Virus". Clinical and Vaccine Immunology. 15 (8): 1214–1221. doi:10.1128/CVI.00120-08. PMC 2519314. PMID 18524884.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Donald J. Benton, Andrea Nans, Lesley J. Calder, Jack Turner, Ursula Neu, Yi Pu Lin, Esther Ketelaars, Nicole L. Kallewaard, Davide Corti, Antonio Lanzavecchia, Steven J. Gamblin, Peter B. Rosenthal, John J. Skehel (Oct 2, 2018) [Sep 17, 2018]. "Hemagglutinin membrane anchor". Proceedings of the National Academy of Sciences of the United States of America. 115 (40): 10112–10117. doi:10.1073/pnas.1810927115. PMC 6176637. PMID 30224494.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Payne, Susan (2017). "Methods to Study Viruses". Viruses. pp. 37–52. doi:10.1016/B978-0-12-803109-4.00004-0. ISBN 978-0-12-803109-4. S2CID 89981392.
- ↑ Ashiba, Hiroki; Fujimaki, Makoto; Awazu, Koichi; Fu, Mengying; Ohki, Yoshimichi; Tanaka, Torahiko; Makishima, Makoto (March 2015). "Hemagglutination detection for blood typing based on waveguide-mode sensors". Sensing and Bio-Sensing Research. 3: 59–64. doi:10.1016/j.sbsr.2014.12.003.
External links
 Media related to Haemagglutinin at Wikimedia Commons
Media related to Haemagglutinin at Wikimedia Commons