Mycobacterium leprae
| Mycobacterium leprae | |
|---|---|
 | |
| Microphotograph of Mycobacterium leprae taken from a skin lesion. The small brick-red rod-shaped cells appear in clusters. Source: CDC | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Actinomycetota |
| Class: | Actinomycetia |
| Order: | Mycobacteriales |
| Family: | Mycobacteriaceae |
| Genus: | Mycobacterium |
| Species: | M. leprae |
| Binomial name | |
| Mycobacterium leprae Hansen, 1874 | |
Mycobacterium leprae (also known as the leprosy bacillus or Hansen's bacillus), is one[lower-alpha 1] of the two species of bacteria that cause Hansen’s disease (leprosy),[1] a chronic but curable infectious disease that damages the peripheral nerves and targets the skin, eyes, nose, and muscles.[2]
It is an acid-fast, Gram-positive, rod shaped bacterium and an obligate intracellular parasite, which means, unlike its relative Mycobacterium tuberculosis, it cannot be grown in cell-free laboratory media.[3] This is likely due to gene deletion and decay that the genome of the species has experienced via reductive evolution, which has caused the bacterium to depend heavily on its host for nutrients and metabolic intermediates.[4] It has a narrow host range and apart from humans, the only other natural hosts are nine-banded armadillo and red squirrels.[5] The bacteria infect mainly macrophages and Schwann cells, and are typically found congregated as a palisade.[6][7]
Mycobacterium leprae was sensitive to dapsone as a treatment alone, but since the 1960's, it has developed resistance against this antibiotic. Currently, a multidrug treatment (MDT) is recommended by the World Health Organization, including dapsone, rifampicin, and clofazimine. The species was discovered in 1873 by the Norwegian physician Gerhard Armauer Hansen, and was the first bacterium to be identified as a cause of disease in humans.[8]
Microbiology
Mycobacterium leprae is an intracellular, pleomorphic, non-sporing, non-motile, acid-fast, pathogenic bacterium.[3] It is an aerobic bacillus (rod-shaped bacterium) with parallel sides and round ends, surrounded by the characteristic waxy coating of mycolic acid unique to mycobacteria. It is Gram-positive by Gram staining, but Mycobacterium leprae was traditionally stained with carbol fuchsin in the Ziehl–Neelsen stain. Because the bacilli are less acid-fast than Mycobacterium tuberculosis (MTB), the Fite-Faraco staining method, which has a lower acid concentration, is used now.[9][10] In size and shape, it closely resembles MTB. The bacteria are found in the granulomatous lesions and are especially numerous in the nodules. This bacteria often occur in large numbers within the lesions of lepromatous leprosy and are usually grouped together as a palisade.[6] By optical microscopy of host cells, Mycobacterium leprae can be found singly or in clumps referred to as "globi", the bacilli can be straight or slightly curved, with a length ranging from 1–8 μm and a diameter of 0.3 μm.[11] The bacteria grow best at 27 to 30°C, making the skin, nasal mucosa and peripheral nerves primary targets for infection by Mycobacterium leprae.[12]
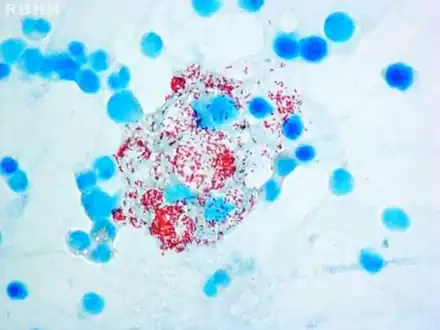 Histiocytes with their abundant cytoplasm filled with acid-fast bacilli (Mycobacterium leprae) in bone marrow
Histiocytes with their abundant cytoplasm filled with acid-fast bacilli (Mycobacterium leprae) in bone marrow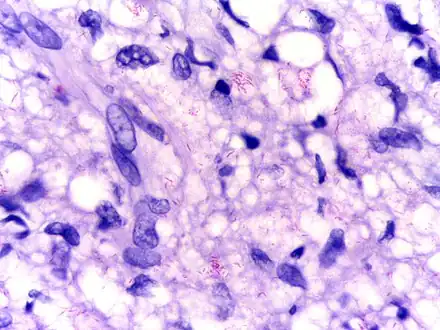 Modified AFB staining in a case of lepromatous leprosy showing numerous rod shaped acid fast bacilli
Modified AFB staining in a case of lepromatous leprosy showing numerous rod shaped acid fast bacilli
Host range
Mycobacterium leprae has a narrow host range and apart from humans the only other hosts are nine-banded armadillos and red squirrels,[5] and armadillos have been implicated as a source of zoonotic leprosy in humans.[13] In the laboratory, mice can be infected and this is a useful animal model.[14]
Cultivation
Efforts to culture the bacteria in vitro remain unsuccessful.[15] Because the organism is an obligate intracellular parasite, it lacks many necessary genes for independent survival, causing difficulty in culturing the organism. The complex and unique cell wall that makes members of the genus Mycobacterium difficult to destroy is also the reason for its extremely slow replication rate. Mycobacterium leprae prefers cool temperatures, slightly acidic microaerophilic conditions, and prefers the use of lipids as an energy source over sugars. The growth conditions needed for Mycobacterium leprae are known, but an exact axenic medium to support the growth of Mycobacterium leprae still has yet to be discovered.[16] Since in vitro cultivation is not generally possible, it has instead been grown in mouse foot pads,[14] and in armadillos due to their low core body temperature.[17][16]
Metabolism
The reductive evolution experienced by the Mycobacterium leprae genome has impaired its metabolic abilities in comparison to other Mycobacterium, specifically in its catabolic pathways. [18]
Catabolism
Mycobacterium leprae’s inability to be grown in axenic media indicates its reliance on nutrients and intermediates from its host.[19] Many of the catabolic pathways present in other Mycobacterium species are compromised, due to the absence of enzymes that play key roles in degradation of nutrients.[19] Mycobacterium leprae has lost the ability to use common carbon sources, such as acetate and galactose, in its central energy metabolism pathways.[4] Additionally, lipid degradation is impaired, with deficits in key lipase enzymes, and other proteins involved in lipolysis.[20] Functional carbon catabolic pathways continue to exist in the species, such as the glycolytic pathway, the pentose phosphate pathway, and the TCA cycle.[19] These deficiencies extensively restricts the microbe's growth to a limited number of carbon sources, such as host-derived intermediates.[4]
Anabolism
Mycobacterium leprae's anabolic pathways have been largely unaffected by its reductive evolution.[18] The species retains its ability for the synthesis of genetic material, such as purines, pyrimidines, nucleotides, and nucleosides, as well as the synthesis of all amino acids, except for methionine and lysine.[19]
Genome
Mycobacterium leprae has the unusual properties among other microbes, such as its lengthy doubling time (ranging from 12–14 days compared with 20 minutes for Escherichia coli), as well as its inability to be cultured in the laboratory.[21][22] Comparing the genome sequence of Mycobacterium leprae with that of MTB offers explanations for these properties, and reveals an extreme case of reductive evolution. Less than half of the genome contains functional genes. It is estimated approximately 2000 genes from Mycobacterium leprae genome has been lost.[23] Gene deletion and decay appear to have eliminated many important metabolic activities, including siderophore production, part of the oxidative and most of the microaerophilic and anaerobic respiratory chains, and numerous catabolic systems and their regulatory circuits.[24] This reductive evolution is largely linked to the organism's development into an obligate intracellular bacterium.[25]
The first genome sequence of a strain of Mycobacterium leprae was completed in 2001, which revealed only half of the genome contains protein-coding regions and the other half consists of pseudogenes and non-coding regions.[23] The genome sequence of a strain originally isolated in Tamil Nadu, India, and designated TN, was completed in 2013. The sequence was obtained by a combined approach, employing automated DNA sequence analysis of selected cosmids and whole-genome 'shotgun' clones. After the finishing process, the genome sequence was found to contain 3,268,203 base pairs (bp), and to have an average G+C content of 57.8%, values much lower than the corresponding values for M. tuberculosis, which are 4,441,529 bp and 65.6% G+C.[26]
Pseudogenes
Many of the genes that were present in the genome of the common ancestor of Mycobacterium leprae and M. tuberculosis have been lost in the Mycobacterium leprae genome.[23][27]Due to Mycobacterium leprae's reliance on a host organism, many of the species' DNA repair functions have been lost, increasing the occurrence of deletion mutations.[25] Because the products supplied by these deleted genes are typically present in the host cells infected by Mycobacterium leprae, the impact that the mutations have on the microbe is minimal, allowing for survival within the host despite its reduced genome.[28] Consequently, Mycobacterium leprae has undergone a dramatic reduction in genome size with the loss of many genes.[4] Over half of the pathogen's genome is now made up by pseudogenes due to the pathogen undergoing what is known as reductive evolution. [4] Among published genomes, Mycobacterium leprae contains the highest number of pseudogens (>1000). [29] Many of the pseudogenes in Mycobacterium leprae arose from insertions of stop codons which may have been caused by sigma factor dysfunction (a protein needed for initiation of transcription in bacteria) or the insertion of transposon- derived repetitive sequences.[30] Some of the Mycobacterium leprae pseudogens expression levels will alter upon infection of macrophages, which suggests that some Mycobacterium leprae pseudogens are not all "decayed" genes, but could also function in infection, intracellular replication, and replication.[29] This genome reduction is not complete.[27] Downsizing from a genome of 4.42 Mbp, such as that of M. tuberculosis, to one of 3.27 Mbp would account for the loss of some 1200 protein-coding sequences.
Essential Enzymes
There are eight essential enzymes for Mycobacterium leprae, and one of them is alanine racemase (alr). This enzyme is significant because it is found in D-alanine—D-alanine ligase and alanine/Aspartate metabolism. Other essential enzymes include putative dTDP4deydrorhamnose 3, 5epimerase (rm1C) which plays an important role in both Nucleotide sugar metabolism and polyketide sugar unit biosynthesis. Petidoglycan biosynthesis also require murG, murF, MurE, murY, murC, and murD, the remaining six essential enzymes for mycobacterium leprae. [31]
Distribution
The bacterium has a global distribution in humans but the highest prevalence is in sub-Saharan Africa, Asia and South America.[32] The geographic occurrences of Mycobacterium leprae include: Angola, Brazil, Central African Republic, the Democratic Republic of Congo, Federated States of Micronesia, India, Kiribati, Madagascar, Nepal, Republic of Marshall Islands, and the United Republic of Tanzania.[33]
Since the introduction of multidrug therapy (MDT) in the 1980s, the prevalence of leprosy cases has declined by 95%.[34] This decline led the World Health Organization (WHO) to declare leprosy eliminated as a public health problem, defined as a prevalence of less than one leprosy patient per 10,000 population.[35] Aside from Mycobacterium leprae transmission from infected humans, environmental sources could also be an important reservoir. Mycobacterium leprae DNA was detected in soil from houses of leprosy patients in Bangladesh, armadillos’ holes in Suriname and habitats of lepromatous red squirrels in the British Isles.[36] One study found numerous reports of leprosy cases with a history of contact with armadillos in the United States.[34] A zoonotic transmission pathway from exposure to armadillos has been proposed, with human patients from a previous study in southeastern United States shown to be infected with the same armadillo-associated Mycobacterium leprae genotype.[37] High rates of Mycobacterium leprae infection were observed in armadillos in the Brazilian state of Pará, and individuals who frequently consumed armadillo meat showed a significantly higher titres of the M. leprae-specific antigen, phenolic glycolipid I (PGL-I) compared with those who did not or ate them less frequently.[38][34]
Evolution
The closest relative to Mycobacterium leprae is Mycobacterium lepromatosis. These species diverged 13.9 million years ago (95% highest posterior density 8.2 million years ago – 21.4 million years ago ) The most recent common ancestor of the extant Mycobacterium leprae strains was calculated to have lived 3,607 years ago (95% highest posterior density 2204–5525 years ago). The estimated substitution rate was 7.67 x 10−9 substitutions per site per year, similar to other bacteria.[39]
A study of genomes isolated from medieval cases estimated the mutation rate to be 6.13 × 10−9. The authors also showed that the leprosy bacillus in the Americas was brought there from Europe.[40] Another study suggests that Mycobacterium leprae originated in East Africa and spread from there to Europe and the Middle East initially before spreading to West Africa and the Americas in the last 500 years.[41]
Almost complete sequences of Mycobacterium leprae from medieval skeletons with osteological lesions suggestive of leprosy from different Europe geographic origins were obtained using DNA capture techniques and high-throughput sequencing. Ancient sequences were compared with those of modern strains from biopsies of leprosy patients representing diverse genotypes and geographic origins, giving new insights in the understanding of its evolution and course through history, phylogeography of the leprosy bacillus, and the disappearance of leprosy from Europe.[40]
Verena J. Schuenemann et al. demonstrated a remarkable genomic conservation during the past 1000 years and a close similarity between modern and ancient strains, suggesting that the sudden decline of leprosy in Europe was not due to a loss of virulence, but due to extraneous factors, such as other infectious diseases, changes in host immunity, or improved social conditions.[40]
Pathogenesis

The incubation period of Mycobacterium leprae ranges from 9 months to 20 years.[43] The bacterium replicates intracellularly inside histiocytes and nerve cells and has two forms. One form is "tuberculoid," which induces a cell-mediated response that limits its growth, and has few detectible bacilli (paucibacillary).[44] Through this form, Mycobacterium leprae multiplies at the site of entry, usually the skin, invading and colonizing Schwann cells. The bacterium then induces T-helper lymphocytes, epithelioid cells, and giant cell infiltration of the skin, causing infected individuals to exhibit large flattened patches with raised and elevated red edges on their skin. These patches have dry, pale, hairless centers, accompanied by a loss of sensation on the skin. The loss of sensation may develop as a result of invasion of the peripheral sensory nerves. The macule at the cutaneous site of entry and the loss of pain sensation are key clinical indications that an individual has a tuberculoid form of leprosy.[45]
The second form of leprosy is the "lepromatous" form, in which the microbes proliferate within the macrophages at the site of entry, and has many detectable bacilli (multibacillary).[44] They also grow within the epithelial tissues of the face and ear lobes. The suppressor T-cells that are induced are numerous, but the epithelioid and giant cells are rare or absent. With cell-mediated immunity impaired, large numbers of Mycobacterium leprae appear in the macrophages and the infected patients develop papules at the entry site, marked by a folding of the skin. Gradual destruction of cutaneous nerves lead to what is referred to as "classic lion face." Extensive penetration by this bacterium may lead to severe body damage; for example the loss of bones, fingers, and toes.[45]
Symptoms of a Mycobacterium leprae infection
The symptoms of a Mycobacterium leprae infection, also known as leprosy, are skin sores that are pale in color, lumps or bumps that do not go away after several weeks or months, nerve damage which can lead to complications with the ability to sense feeling in the arms and legs as well as muscle weakness. Symptoms usually take 3–5 years from being exposed to manifest within the body. However, some individuals do not begin to show symptoms until 20 years after exposure to the disease. This long incubation period makes the ability to properly be able to diagnose when an individual came into contact with the disease very difficult.[46]
In armadillos, Mycobacterium leprae causes a disseminated infection with similar structural and pathological changes in tissues and nerves.[47]
In squirrels, according the to Veterinary Pathology Unit of the University of Edinburgh, " The disease is unmistakeable: there is gross swelling and loss of hair around the snout, lips, eyelids, ears, genitalia and sometimes feet and lower limbs. This bare skin has a “shiny” appearance. The squirrel is usually in generally poor body condition and may have a heavy burden of parasites like fleas, ticks and mites."[48]
Treatment
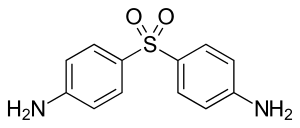
The mycolic acids in the bacteria's cell walls afford resistance to many antibiotics and are a major virulence factor.[49] Multidrug therapy (MDT) was recommended by WHO Expert Committee in 1984, and became the standard leprosy treatment. MDT has been supplied by WHO for free since 1995 to endemic countries. MDT is used to treat leprosy because treatment of leprosy with one drug (monotherapy) can result in drug resistance. The drug combination used in MDT will depend on the classification of the disease. WHO recommends patients with multibacillary leprosy use a combination of Rifampicin, Clofazimine, and Dapsone for 12 months. WHO recommends patients with paulibacilalry leprosy use combination of Rifampicin and Dapsone for a duration of 6 months.[50] Antibiotics must be taken regularly until treatment is complete because Mycobacterium leprae can become drug resistant.[51] Effectiveness of the treatment can be determined with the use of an acid-fast stain of Mycobacterium leprae from a skin smear to estimate the number of bacilli still present in the patient.[52]
The number of reported cases of leprosy annually is around 250,000 cases indicating that the chain of transmission has yet to be broken despite the use of MDT leading to a 90% reduction in the prevalence rate of leprosy. This makes it very evident that control of the disease is not yet where it needs to be calling for the need in continued research towards treatment and control. [4]
A preventive measure of Mycobacterium leprae is to avoid close contact with infectious people who are untreated.[53] Blindness, crippling of the hands and feet, and paralysis are all effects of nerve damage associated with untreated M. leprae. Treatment does not reverse the nerve damage done, which is why early treatment is needed.[51] The Bacillus Calmette–Guérin vaccine offers a variable amount of protection against leprosy in addition to its main target of tuberculosis.[54]
Targets of antibiotics
Dapsone competitively inhibits the enzyme dihydropteroate synthase (DHPS) resulting in decreases the production of tetrahydrofolate, which is an essential component of nucleic acid biosynthesis in M. leprae. Rifampin will interrupt binding of the β-subunit of the DNA-dependent RNA polymerase, which will uncouple mRNA production and results in cell death. Clofazimine's mechanisms are not fully understood regarding Mycobacterium leprae, but the drug's binding appear to occur at base sequences with guanine, which may explain why clofazimine has a preference for G+C rich genomes of mycobacteria over human DNA. The binding of clofazimine to mycobacterial DNA can has been proven as weakly bactericidal against Mycobacterium leprae in mice, which is why it is not suitable for single drug therapy for leprosy. Out of the three main drugs rifampin is more bactericidal than either dapsone or clofazimine. [55]
Potential antibiotic targets
It is important to find new targets for antibiotics due to increasing resistance. Mycobacterium leprae has six essential enzymes murC, murD, murE, murF, murG, and murY that are all essential for peptidoglycan biosynthesis in M. leprae. These enzymes and peptidoglycan biosynthesis are potential targets for antibiotics. By targeting these enzymes, which catalyze additions of short polypeptide chains, synthesis of the bacterial cell wall can be prevented.[56]
Antibiotic resistance
Resistance to antibiotics is seen in around 10% of new cases of leprosy and in around 15% of relapsed cases.[57] Drug resistance in Mycobacterium leprae is thought to be from genetic alterations in the antibiotic targets and a reduction in cell wall permeability.[58] Compared to the amount of efflux pumps in M. tuberculosis, Mycobacterium leprae contains about half as many. The efflux pumps contributing to drug resistance and virulence in M. tuberculosis have been retained throughout the genome reductive evolution that Mycobacterium leprae underwent.[58]
Discovery

Mycobacterium leprae was discovered in 1873 by the Norwegian physician Gerhard Armauer Hansen (1841–1912), and was the first bacterium to be identified as a cause of disease in humans.[8]
It was confirmed to be a bacterium by Albert Ludwig Sigesmund Neisser who argued with Hansen over priority for the discovery.[59]
Hansen's attempts to infect animals with the bacteria were unsuccessful and he was dismissed from his post at the Leprosy Hospital in Bergen and was banned from practising medicine when, in 1879, he injected, without consent, tissue from a person with lepromatous leprosy into the eye of 33-year old Kari Nielsdatter who had the milder tuberculoid form of the infection.[60]
The case had little effect on Hansen’s professional reputation, and he continued with his research.[61]
Notes
- ↑ The other is Mycobacterium lepromatosis.
References
- ↑ Serrano-Coll H, Cardona-Castro N (June 2022). "Neuropathic ulcers in leprosy: clinical features, diagnosis and treatment". Journal of Wound Care. 31 (Sup6): S32–S40. doi:10.12968/jowc.2022.31.Sup6.S32. PMID 35678776. S2CID 249521365.
- ↑ "Mycobacterium Leprae, the Cause of Leprosy". Microbiology Society. August 27, 2014. Archived from the original on November 12, 2019. Retrieved November 12, 2019.
- 1 2 McMurray DN (1996). "Mycobacteria and Nocardia.". In Baron S., et al. (eds.). Baron's Medical Microbiology (4th ed.). University of Texas Medical Branch. ISBN 978-0-9631172-1-2. PMID 21413269. Archived from the original on February 12, 2009. Retrieved September 5, 2017.
- 1 2 3 4 5 6 Singh P, Cole ST (January 2011). "Mycobacterium leprae: genes, pseudogenes and genetic diversity". Future Microbiology. 6 (1): 57–71. doi:10.2217/fmb.10.153. PMC 3076554. PMID 21162636.
- 1 2 Sharma R, Singh P, Pena M, Subramanian R, Chouljenko V, Kim J, et al. (August 2018). "Differential growth of Mycobacterium leprae strains (SNP genotypes) in armadillos". Infection, Genetics and Evolution. 62: 20–26. doi:10.1016/j.meegid.2018.04.017. PMID 29665434.
- 1 2 "Microbiology of M.leprae". World Health Organization. Archived from the original on June 8, 2013.
- ↑ Leal-Calvo T, Martins BL, Bertoluci DF, Rosa PS, de Camargo RM, Germano GV, Brito de Souza VN, Pereira Latini AC, Moraes MO (2021). "Large-Scale Gene Expression Signatures Reveal a Microbicidal Pattern of Activation in Mycobacterium leprae-Infected Monocyte-Derived Macrophages With Low Multiplicity of Infection". Frontiers in Immunology. 12: 647832. doi:10.3389/fimmu.2021.647832. PMC 8085500. PMID 33936067.
- 1 2 Hansen GA (1874). Undersøgelser Angående Spedalskhedens Årsager [Investigations concerning the etiology of leprosy] (in Danish). OCLC 969496922. Archived from the original on September 29, 2022. Retrieved April 9, 2021.
{{cite book}}: CS1 maint: unrecognized language (link) - ↑ Kalagarla S, Alluri R, Saka S, Godha V, Undavalli N, Kolalapudi SA (May 2022). "Efficacy of fluorescent microscopy versus modified Fite-Faraco stain in skin biopsy specimens of leprosy cases - a comparative study". International Journal of Dermatology. 61 (5): 595–599. doi:10.1111/ijd.16046. PMID 35061916. S2CID 246165881.
- ↑ Froes LA, Sotto MN, Trindade MA (2022). "Leprosy: clinical and immunopathological characteristics". Anais Brasileiros de Dermatologia. 97 (3): 338–347. doi:10.1016/j.abd.2021.08.006. PMC 9133310. PMID 35379512.
- ↑ Shinnick TM (2006). "Mycobacterium leprae". In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds.). The Prokaryotes. Springer. pp. 934–44. doi:10.1007/0-387-30743-5_35. ISBN 978-0-387-25493-7. Archived from the original on September 29, 2020. Retrieved July 14, 2019.
- ↑ Gorbach SL, Bartlett JG, Blacklow NR (1992). Infectious diseases. Philadelphia: Saunders. p. 1882. ISBN 0-7216-4168-7. OCLC 22346573.
- ↑ Walsh GP, Meyers WM, Binford CH, Gormus BJ, Baskin GB, Wolf RH, Gerone PJ (1988). "Leprosy as a zoonosis: an update". Acta Leprologica. 6 (1): 51–60. PMID 3051854.
- 1 2 Adams LB (May 2021). "Susceptibility and resistance in leprosy: Studies in the mouse model". Immunological Reviews. 301 (1): 157–174. doi:10.1111/imr.12960. PMC 8252540. PMID 33660297.
- ↑ Gillis TP (2015). "Mycobacterium leprae". Molecular Medical Microbiology (Second Edition): 1655–1668. doi:10.1016/B978-0-12-397169-2.00093-7. ISBN 9780123971692.
- 1 2 Lahiri R, Adams LB. (2016) "Cultivation and viability determination of Mycobacterium leprae", Chapter 5.3. In Scollard DM, Gillis TP (ed), International textbook of leprosy, Archived January 9, 2023, at the Wayback Machine.
- ↑ Sharma R, Singh P, Loughry WJ, Lockhart JM, Inman WB, Duthie MS, et al. (December 2015). "Zoonotic Leprosy in the Southeastern United States". Emerging Infectious Diseases. 21 (12): 2127–2134. doi:10.3201/eid2112.150501. PMC 4672434. PMID 26583204.
- 1 2 Borah K, Girardi KD, Mendum TA, Lery LM, Beste DJ, Lara FA, et al. (December 2019). Boyle JP (ed.). "Intracellular Mycobacterium leprae Utilizes Host Glucose as a Carbon Source in Schwann Cells". mBio. 10 (6). doi:10.1128/mBio.02351-19. PMC 6918074. PMID 31848273.
- 1 2 3 4 Borah K, Kearney JL, Banerjee R, Vats P, Wu H, Dahale S, et al. (July 2020). Borlee BR (ed.). "GSMN-ML- a genome scale metabolic network reconstruction of the obligate human pathogen Mycobacterium leprae". PLOS Neglected Tropical Diseases. 14 (7): e0007871. doi:10.1371/journal.pntd.0007871. PMC 7365477. PMID 32628669.
- ↑ Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, et al. (February 2001). "Massive gene decay in the leprosy bacillus". Nature. 409 (6823): 1007–1011. Bibcode:2001Natur.409.1007C. doi:10.1038/35059006. PMID 11234002. S2CID 4307207.
- ↑ "Mycobacterium leprae". Archived from the original on October 31, 2021. Retrieved October 31, 2021.
- ↑ Truman RW, Krahenbuhl JL (March 2001). "Viable M. leprae as a research reagent". International Journal of Leprosy and Other Mycobacterial Diseases. 69 (1): 1–12. PMID 11480310.
- 1 2 3 Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, et al. (February 2001). "Massive gene decay in the leprosy bacillus". Nature. 409 (6823): 1007–1011. Bibcode:2001Natur.409.1007C. doi:10.1038/35059006. PMID 11234002. S2CID 4307207.
- ↑ Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. (June 1998). "Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence". Nature. 393 (6685): 537–544. Bibcode:1998Natur.393..537C. doi:10.1038/31159. PMID 9634230.
- 1 2 Chavarro-Portillo B, Soto CY, Guerrero MI (September 2019). "Mycobacterium leprae's evolution and environmental adaptation". Acta Tropica. 197: 105041. doi:10.1016/j.actatropica.2019.105041. PMID 31152726. S2CID 173188912.
- ↑ Narayanan S, Deshpande U (June 2013). "Whole-Genome Sequences of Four Clinical Isolates of Mycobacterium tuberculosis from Tamil Nadu, South India". Genome Announcements. 1 (3): e00186–13. doi:10.1128/genomeA.00186-13. PMC 3707582. PMID 23788533.
- 1 2 Chavarro-Portillo B, Soto CY, Guerrero MI (September 2019). "Mycobacterium leprae's evolution and environmental adaptation". Acta Tropica. 197: 105041. doi:10.1016/j.actatropica.2019.105041. PMID 31152726. S2CID 173188912.
- ↑ Vissa VD, Brennan PJ (August 3, 2001). "The genome of Mycobacterium leprae: a minimal mycobacterial gene set". Genome Biology. 2 (8): REVIEWS1023. doi:10.1186/gb-2001-2-8-reviews1023. PMC 138955. PMID 11532219.
- 1 2 Nakamura K, Akama T, Bang PD, Sekimura S, Tanigawa K, Wu H, et al. (September 2009). "Detection of RNA expression from pseudogenes and non-coding genomic regions of Mycobacterium leprae". Microbial Pathogenesis. 47 (3): 183–187. doi:10.1016/j.micpath.2009.06.006. PMID 19555754.
- ↑ Akama T, Suzuki K, Tanigawa K, et al. Whole-genome tiling array analysis of Mycobacterium leprae RNA reveals high expression of pseudogenes and noncoding regions. J Bacteriol. 2009;191(10):3321–3327. DOI:10.1128/JB.00120-09
- ↑ Shanmugam, Anusuya; Natarajan, Jeyakumar (March 31, 2010). "Computational genome analyses of metabolic enzymes in Mycobacterium leprae for drug target identification". Bioinformation. 4 (9): 392–395. doi:10.6026/97320630004392. ISSN 0973-2063. PMC 2951640. PMID 20975887.
- ↑ Reibel F, Chauffour A, Brossier F, Jarlier V, Cambau E, Aubry A (2015). "New Insights into the Geographic Distribution of Mycobacterium leprae SNP Genotypes Determined for Isolates from Leprosy Cases Diagnosed in Metropolitan France and French Territories". PLOS Neglected Tropical Diseases. 9 (10): e0004141. doi:10.1371/journal.pntd.0004141. PMC 4595418. PMID 26441080.
- ↑ "Risk of Exposure | Hansen's Disease (Leprosy) | CDC". www.cdc.gov. Archived from the original on July 19, 2017. Retrieved November 17, 2015.
- 1 2 3 Hambridge T, Nanjan Chandran SL, Geluk A, Saunderson P, Richardus JH (May 2021). "Mycobacterium leprae transmission characteristics during the declining stages of leprosy incidence: A systematic review". PLOS Neglected Tropical Diseases. 15 (5): e0009436. doi:10.1371/journal.pntd.0009436. PMC 8186771. PMID 34038422.
- ↑ Smith WC, van Brakel W, Gillis T, Saunderson P, Richardus JH (April 2015). "The missing millions: a threat to the elimination of leprosy". PLOS Neglected Tropical Diseases. 9 (4): e0003658. doi:10.1371/journal.pntd.0003658. PMC 4408099. PMID 25905706.
- ↑ Tió-Coma M, Wijnands T, Pierneef L, Schilling AK, Alam K, Roy JC, Faber WR, Menke H, Pieters T, Stevenson K, Richardus JH, Geluk A (February 2019). "Detection of Mycobacterium leprae DNA in soil: multiple needles in the haystack". Scientific Reports. 9 (1): 3165. Bibcode:2019NatSR...9.3165T. doi:10.1038/s41598-019-39746-6. PMC 6395756. PMID 30816338.
- ↑ Sharma R, Singh P, Loughry WJ, Lockhart JM, Inman WB, Duthie MS, Pena MT, Marcos LA, Scollard DM, Cole ST, Truman RW (December 2015). "Zoonotic Leprosy in the Southeastern United States". Emerging Infectious Diseases. 21 (12): 2127–34. doi:10.3201/eid2112.150501. PMC 4672434. PMID 26583204.
- ↑ da Silva MB, Portela JM, Li W, Jackson M, Gonzalez-Juarrero M, Hidalgo AS, Belisle JT, Bouth RC, Gobbo AR, Barreto JG, Minervino AH, Cole ST, Avanzi C, Busso P, Frade MA, Geluk A, Salgado CG, Spencer JS (June 2018). "Evidence of zoonotic leprosy in Pará, Brazilian Amazon, and risks associated with human contact or consumption of armadillos". PLOS Neglected Tropical Diseases. 12 (6): e0006532. doi:10.1371/journal.pntd.0006532. PMC 6023134. PMID 29953440.
- ↑ Singh P, Benjak A, Schuenemann VJ, Herbig A, Avanzi C, Busso P, et al. (April 2015). "Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis". Proceedings of the National Academy of Sciences of the United States of America. 112 (14): 4459–4464. Bibcode:2015PNAS..112.4459S. doi:10.1073/pnas.1421504112. PMC 4394283. PMID 25831531.
- 1 2 3 Schuenemann VJ, Singh P, Mendum TA, Krause-Kyora B, Jäger G, Bos KI, et al. (July 2013). "Genome-wide comparison of medieval and modern Mycobacterium leprae". Science. 341 (6142): 179–183. Bibcode:2013Sci...341..179S. doi:10.1126/science.1238286. PMID 23765279. S2CID 22760148.
- ↑ Monot M, Honoré N, Garnier T, Araoz R, Coppée JY, Lacroix C, et al. (May 2005). "On the origin of leprosy". Science. 308 (5724): 1040–1042. doi:10.1126/science/1109759. PMID 15894530. S2CID 86109194. Archived from the original on August 3, 2020. Retrieved July 14, 2019.
- ↑ Cabral N, de Figueiredo V, Gandini M, de Souza CF, Medeiros RA, Lery LM, Lara FA, de Macedo CS, Pessolani MC, Pereira GM (2022). "Modulation of the Response to Mycobacterium leprae and Pathogenesis of Leprosy". Frontiers in Microbiology. 13: 918009. doi:10.3389/fmicb.2022.918009. PMC 9201476. PMID 35722339.
- ↑ "Leprosy (Hansen's disease) – Blue Book – Department of Health and Human services, Victoria, Australia". ideas.health.vic.gov.au. Archived from the original on November 8, 2015. Retrieved November 17, 2015.
- 1 2 Hess S, Rambukkana A (July 2019). Cossart P, Roy CR, Sansonetti P (eds.). "Cell Biology of Intracellular Adaptation of Mycobacterium leprae in the Peripheral Nervous System". Microbiology Spectrum. 7 (4): 7.4.13. doi:10.1128/microbiolspec.BAI-0020-2019. PMC 6700727. PMID 31322104.
- 1 2 Frade MA, Coltro PS, Filho FB, Horácio GS, Neto AA, da Silva VZ, et al. (2022). "Lucio's phenomenon: A systematic literature review of definition, clinical features, histopathogenesis and management". Indian Journal of Dermatology, Venereology and Leprology. 88 (4): 464–477. doi:10.25259/IJDVL_909_19. PMID 34672479. S2CID 239051859.
- ↑ "Leprosy Overview". WebMD. Archived from the original on May 27, 2018. Retrieved November 14, 2019.
- ↑ Oliveira IV, Deps PD, Antunes JM (September 2019). "Armadillos and leprosy: from infection to biological model". Revista do Instituto de Medicina Tropical de Sao Paulo. 61: e44. doi:10.1590/S1678-9946201961044. PMC 6746198. PMID 31531622.
- ↑ "Red squirrel leprosy, The University of Edinburgh" (PDF). Archived (PDF) from the original on December 31, 2021. Retrieved January 1, 2023.
- ↑ Biegas KJ, Swarts BM (December 2021). "Chemical probes for tagging mycobacterial lipids". Current Opinion in Chemical Biology. 65: 57–65. doi:10.1016/j.cbpa.2021.05.009. PMID 34216933.
- ↑ World Health Organization. "Guidelines for the diagnosis, treatment and prevention of leprosy." (2018).
- 1 2 "Diagnosis and Treatment | Hansen's Disease (Leprosy) | CDC". www.cdc.gov. November 2, 2018. Archived from the original on November 8, 2019. Retrieved November 12, 2019.
- ↑ Lahiri R, Adams LB. 18 September 2016, posting date. Cultivation and viability determination of Mycobacterium leprae, Chapter 5.3. In Scollard DM, Gillis TP (ed), International textbook of leprosy. www.internationaltextbookofleprosy.org.
- ↑ "Leprosy: MedlinePlus Medical Encyclopedia". www.nlm.nih.gov. Archived from the original on October 6, 2015. Retrieved November 17, 2015.
- ↑ Duthie MS, Gillis TP, Reed SG (November 2011). "Advances and hurdles on the way toward a leprosy vaccine". Human Vaccines. 7 (11): 1172–1183. doi:10.4161/hv.7.11.16848. PMC 3323495. PMID 22048122.
- ↑ Cambau E, Williams DL. 25 January 2019, posting date. Anti-leprosy drugs: Modes of action and mechanisms of resistance in Mycobacterium leprae, Chapter 5.2. In Scollard DM, Gillis TP (ed), International textbook of leprosy. www.internationaltextbookofleprosy.org.
- ↑ Shanmugam, Anusuya; Natarajan, Jeyakumar (2010). "Computational genome analyses of metabolic enzymes in Mycobacterium leprae for drug target identification". Bioinformation. 4 (9): 392–395. doi:10.6026/97320630004392. PMC 2951640. PMID 20975887. Archived from the original on November 17, 2022. Retrieved November 17, 2022.
- ↑ Li X, Li G, Yang J, Jin G, Shao Y, Li Y, Wei P, Zhang L (October 2022). "Drug Resistance (Dapsone, Rifampicin, Ofloxacin) and Resistance-Related Gene Mutation Features in Leprosy Patients: A Systematic Review and Meta-Analysis". International Journal of Molecular Sciences. 23 (20): 12443. doi:10.3390/ijms232012443. PMC 9604410. PMID 36293307.
{{cite journal}}: Check|pmc=value (help) - 1 2 Machado D, Lecorche E, Mougari F, Cambau E, Viveiros M (2018). "Insights on Mycobacterium leprae Efflux Pumps and Their Implications in Drug Resistance and Virulence". Frontiers in Microbiology. 9: 3072. doi:10.3389/fmicb.2018.03072. PMC 6300501. PMID 30619157.
- ↑ Bieliaieva O, Uvarkina O, Lysanets Y, Morokhovets H, Honcharova Y, Melaschenko M (December 2020). "Gerhard Hansen Vs. Albert Neisser: Priority For the invention of Mycobacterium Leprae and problems of bioethics". Georgian Medical News (309): 156–1161. PMID 33526747.
- ↑ Dobson MJ (2008). Disease. Englewood Cliffs, N.J: Quercus. p. 25. ISBN 978-1-84724-399-7.
- ↑ Lanska DJ (2015). "Armauer Hansen: The Controversy Surrounding his Unethical Human-to-Human Leprosy Transmission Experiment". Archived from the original on December 2, 2022. Retrieved October 30, 2020.
External links
- The genome of Mycobacterium leprae Archived January 7, 2011, at the Wayback Machine
- "Mycobacterium leprae". NCBI Taxonomy Browser. 1769. Archived from the original on June 2, 2019. Retrieved January 1, 2023.