Opisthokont
| Opisthokont Temporal range: | |
|---|---|
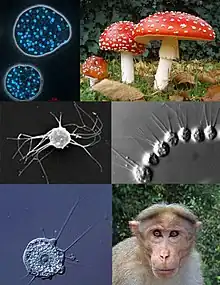 | |
| Clockwise, from top left: Abeoforma whisleri (Mesomycetozoea); Amanita muscaria (Fungi); Desmarella moniliformis (Choanoflagellatea); Bonnet Macaque (Metazoa); Nuclearia thermophila (Nucleariida); Ministeria vibrans (Filasterea) | |
| Scientific classification | |
| (unranked): | Unikonta |
| (unranked): | Obazoa |
| (unranked): | Opisthokonta Copeland 1956,[2] emend. Cavalier-Smith 1987,[3] emend. Adl et al., 2005[4] |
| Subgroups | |
The opisthokonts (from Ancient Greek ὀπίσθιος (opísthios) 'rear, posterior', and κοντός (kontós) 'pole, i.e. flagellum') are a broad group of eukaryotes, including both the animal and fungus kingdoms.[5] The opisthokonts, previously called the "Fungi/Metazoa group",[6] are generally recognized as a clade. Opisthokonts together with Apusomonadida and Breviata comprise the larger clade Obazoa.[7][8][9][10][11]
Flagella and other characteristics
A common characteristic of opisthokonts is that flagellate cells, such as the sperm of most animals and the spores of the chytrid fungi, propel themselves with a single posterior flagellum. It is this feature that gives the group its name. In contrast, flagellate cells in other eukaryote groups propel themselves with one or more anterior flagella. However, in some opisthokont groups, including most of the fungi, flagellate cells have been lost.[7]
Opisthokont characteristics include synthesis of extracellular chitin in exoskeleton, cyst/spore wall, or cell wall of filamentous growth and hyphae; the extracellular digestion of substrates with osmotrophic absorption of nutrients; and other cell biosynthetic and metabolic pathways. Genera at the base of each clade are amoeboid and phagotrophic.[12]
History
The close relationship between animals and fungi was suggested by Thomas Cavalier-Smith in 1987,[3] who used the informal name opisthokonta (the formal name has been used for the chytrids by Copeland in 1956), and was supported by later genetic studies.[13]
Early phylogenies placed fungi near the plants and other groups that have mitochondria with flat cristae, but this character varies. More recently, it has been said that holozoa (animals) and holomycota (fungi) are much more closely related to each other than either is to plants, because opisthokonts have a triple fusion of carbamoyl phosphate synthetase, dihydroorotase, and aspartate carbamoyltransferase that is not present in plants, and plants have a fusion of thymidylate synthase and dihydrofolate reductase not present in the opisthokonts. Animals and fungi are also more closely related to amoebas than to plants, and plants are more closely related to the SAR supergroup of protists than to animals or fungi. Animals and fungi are both heterotrophs, unlike plants, and while fungi are sessile like plants, there are also sessile animals.
Cavalier-Smith and Stechmann argue that the uniciliate eukaryotes such as opisthokonts and Amoebozoa, collectively called unikonts, split off from the other biciliate eukaryotes, called bikonts, shortly after they evolved.[14]
Taxonomy
Opisthokonts are divided into Holomycota or Nucletmycea (fungi and all organisms more closely related to fungi than to animals) and Holozoa (animals and all organisms more closely related to animals than to fungi); no opisthokonts basal to the Holomycota/Holozoa split have yet been identified. The Opisthokonts was largely resolved by Torriella et al.[15] Holomycota and Holozoa are composed of the following groups.
- Holomycota (Fungus-like)
- Fungi
- Includes:
- chytrids (flagellated, zoosporic fungi)
- Fonticula[16] (more recent work considers this to be part of Cristidiscoidea, a sister group to the fungi)
- Hyaloraphidium (previously thought to be a green alga, now considered a fungus)
- microsporidia (previously thought to be apicomplexia)
- Nucleariida (more recent work considers this to be part of Cristidiscoidea, a sister group to the fungi)
- Excludes:
- labyrinthulomycetes (slime nets) (now included in the SAR supergroup)
- myxomycetes (now included in amoebozoans)
- oomycetes (water molds) (now included in the SAR supergroup)
- Includes:
- Rozellida (placement uncertain)
- Fungi
- Holozoa (Animal-like)
- Corallochytrium (formerly considered a Heterokont)
- Filozoa
- Animalia (including myxozoa)
- Choanoflagellata (flagellates formerly included in protozoa)
- Filasterea
- Mesomycetozoea
- Amoebidiales (formerly considered trichomycetes)
- Dermocystida (formerly considered parasitic fungi or sporozoans)
- Eccrinales (formerly considered fungi)
- Ichthyophonida (formerly considered parasitic fungi incertae sedis)
Phylogeny
The choanoflagellates have a circular mitochondrial DNA genome with long intergenic regions. This is four times as large as animal mitochondrial genomes and contains twice as many protein coding genes.
Corallochytrium seem likely to be more closely related to the fungi than to the animals on the basis of the presence of ergosterol in their membranes and being capable of synthesis of lysine via the AAA pathway.
The ichthyosporeans have a two amino acid deletion in their EEF1A1 gene that is considered characteristic of fungi.
The ichthyosporean genome is >200 kilobase pairs in length and consists of several hundred linear chromosomes that share elaborate terminal-specific sequence patterns.
In the following phylogenetic tree it is indicated how many millions of years ago (Mya) the clades diverged into newer clades. The holomycota tree is following Tedersoo et al.[17]
| Eukaryotes |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1850 mya |
One view of the great kingdoms and their stem groups. Phylogeny based on Steenkamp et al 2005,[7] and Eichinger et al, 2005.[18]
Gallery
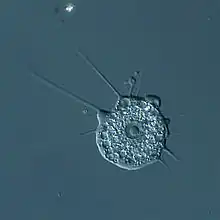
 Rozella sp. (Rozellida)
Rozella sp. (Rozellida) Microsporidian spore (Microsporidia)
Microsporidian spore (Microsporidia) Chytrid (flagellated fungus)
Chytrid (flagellated fungus)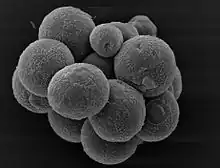 Sphaeroforma sp. (Mesomycetozoea)
Sphaeroforma sp. (Mesomycetozoea)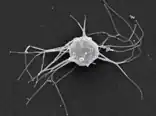 Ministeria sp. (Filasterea)
Ministeria sp. (Filasterea)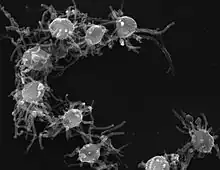 Capsaspora sp. (Filasterea)
Capsaspora sp. (Filasterea)
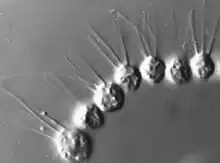 Desmarella sp. colony (Choanoflagellatea)
Desmarella sp. colony (Choanoflagellatea) Two opisthokonts: a human (Metazoa) and a mushroom (Fungi)
Two opisthokonts: a human (Metazoa) and a mushroom (Fungi)
References
- ↑ Loron, Corentin C.; François, Camille; Rainbird, Robert H.; Turner, Elizabeth C.; Borensztajn, Stephen; Javaux, Emmanuelle J. (May 22, 2019). "Early fungi from the Proterozoic era in Arctic Canada". Nature. 570 (7760): 232–235. Bibcode:2019Natur.570..232L. doi:10.1038/s41586-019-1217-0. PMID 31118507. S2CID 162180486.
- ↑ Copeland, H. F. (1956). The Classification of Lower Organisms. Palo Alto: Pacific Books.
- 1 2 Cavalier-Smith, T. (1987). "The origin of fungi and pseudofungi". In Rayner, Alan D. M. (ed.). Evolutionary biology of Fungi. Cambridge: Cambridge University Press. pp. 339–353. ISBN 0-521-33050-5.
- ↑ Adl, S.M.; et al. (September–October 2005). "The new higher level classification of eukaryotes with emphasis on the taxonomy of protists". Journal of Eukaryotic Microbiology. 52 (5): 399–451. doi:10.1111/j.1550-7408.2005.00053.x. PMID 16248873.
- ↑ Shalchian-Tabrizi K, Minge MA, Espelund M, et al. (7 May 2008). Aramayo R (ed.). "Multigene phylogeny of choanozoa and the origin of animals". PLOS ONE. 3 (5): e2098. Bibcode:2008PLoSO...3.2098S. doi:10.1371/journal.pone.0002098. PMC 2346548. PMID 18461162.
- ↑ "Fungi/Metazoa group". Retrieved 2009-03-08.
- 1 2 3 Steenkamp, E.T.; Wright, J.; Baldauf, S.L. (January 2006). "The protistan origins of animals and fungi". Molecular Biology and Evolution. 23 (1): 93–106. doi:10.1093/molbev/msj011. PMID 16151185.
- ↑ Huang, Jinling; Xu, Ying; Gogarten, Johann Peter (November 2005). "The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic". Molecular Biology and Evolution. 22 (11): 2142–2146. doi:10.1093/molbev/msi221. PMID 16049196.
- ↑ Laura Wegener Parfrey; Erika Barbero; Elyse Lasser; Micah Dunthorn; Debashish Bhattacharya; David J Patterson; Laura A Katz (December 2006). "Evaluating support for the current classification of eukaryotic diversity". PLOS Genetics. 2 (12): e220. doi:10.1371/JOURNAL.PGEN.0020220. ISSN 1553-7390. PMC 1713255. PMID 17194223. Wikidata Q21090155.
- ↑ Torruella, Guifré; et al. (February 2012). "Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains". Molecular Biology and Evolution. 29 (2): 531–544. doi:10.1093/molbev/msr185. PMC 3350318. PMID 21771718.
- ↑ Eme, Laura; Sharpe, Susan C.; Brown, Matthew W.; Roger, Andrew J. (August 2014). "On the Age of Eukaryotes: Evaluating Evidence from Fossils and Molecular Clocks". Cold Spring Harbor Perspectives in Biology. 6 (8): a016139. doi:10.1101/cshperspect.a016139. ISSN 1943-0264. PMC 4107988. PMID 25085908.
- ↑ Adl, Sina M.; Bass, David; Lane, Christopher E.; Lukeš, Julius; Schoch, Conrad L.; Smirnov, Alexey; Agatha, Sabine; Berney, Cedric; Brown, Matthew W. (2018-09-26). "Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes". Journal of Eukaryotic Microbiology. 66 (1): 4–119. doi:10.1111/jeu.12691. ISSN 1066-5234. PMC 6492006. PMID 30257078.
- ↑ Wainright, P.O.; Hinkle, G.; Sogin, M.L.; Stickel, S.K. (April 1993). "Monophyletic origins of the metazoa: an evolutionary link with fungi". Science. 260 (5106): 340–342. Bibcode:1993Sci...260..340W. doi:10.1126/science.8469985. PMID 8469985. S2CID 27373608.
- ↑ Stechmann, A.; Cavalier-Smith, T. (5 July 2002). "Rooting the eukaryote tree by using a derived gene fusion". Science. 297 (5578): 89–91. Bibcode:2002Sci...297...89S. doi:10.1126/science.1071196. PMID 12098695. S2CID 21064445.
- ↑ Torruella, Guifré; Mendoza, Alex de; Grau-Bové, Xavier; Antó, Meritxell; Chaplin, Mark A.; Campo, Javier del; Eme, Laura; Pérez-Cordón, Gregorio; Whipps, Christopher M. (2015). "Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi". Current Biology. 25 (18): 2404–2410. doi:10.1016/j.cub.2015.07.053. PMID 26365255.
- ↑ Matthew W. Brown, Frederick W. Spiegel and Jeffrey D. Silberman (2009), "Phylogeny of the "Forgotten" Cellular Slime Mold, Fonticula alba, Reveals a Key Evolutionary Branch within Opisthokonta", Molecular Biology and Evolution, 26 (12): 2699–2709, doi:10.1093/molbev/msp185, PMID 19692665
- ↑ Tedersoo, Leho; Sánchez-Ramírez, Santiago; Kõljalg, Urmas; Bahram, Mohammad; Döring, Markus; Schigel, Dmitry; May, Tom; Ryberg, Martin; Abarenkov, Kessy (2018). "High-level classification of the Fungi and a tool for evolutionary ecological analyses". Fungal Diversity. 90 (1): 135–159. doi:10.1007/s13225-018-0401-0. ISSN 1560-2745.
- ↑ Eichinger, L.; Pachebat, J. A.; Glöckner, G.; Rajandream, M. A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; et al. (2005). "The genome of the social amoeba Dictyostelium discoideum". Nature. 435 (7038): 43–57. Bibcode:2005Natur.435...43E. doi:10.1038/nature03481. PMC 1352341. PMID 15875012.
External links
| Wikispecies has information related to Opisthokonta. |
| Wikimedia Commons has media related to Opisthokonta. |
.jpg.webp)

_(cropped).jpg.webp)