Protein–ligand complex
A protein–ligand complex is a complex of a protein bound with a ligand[2] that is formed following molecular recognition between proteins that interact with each other or with various other molecules. Formation of a protein-ligand complex is based on molecular recognition between biological macromolecules and ligands, where ligand means any molecule that binds the protein with high affinity and specificity. Molecular recognition is not a process by itself since it is part of a functionally important mechanism involving the essential elements of life like in self-replication, metabolism, and information processing. For example DNA-replication depends on recognition and binding of DNA double helix by helicase, DNA single strand by DNA-polymerase and DNA segments by ligase. Molecular recognition depends on affinity and specificity. Specificity means that proteins distinguish the highly specific binding partner from less specific partners and affinity allows the specific partner with high affinity to remain bound even if there are high concentrations of less specific partners with lower affinity.[3]
Interactions
The protein-ligand complex is a reversible non-covalent interaction between two biological (macro)molecules. In non-covalent interactions there is no sharing of electrons like in covalent interactions or bonds. Non-covalent binding may depend on hydrogen bonds, hydrophobic forces, van der Waals forces, π-π interactions, electrostatic interactions in which no electrons are shared between the two or more involved molecules.[4] The molecules (protein and ligand) recognize each other also by stereospecificity i.e. by the form of the two molecules. Because of this real discriminative if not 'cognitive' property, Werner Loewenstein uses the term 'cognitive demon' or molecular demon referring to Maxwell's demon, the famous thought experiment. In fact, the proteins that form complexes are able to pick a substrate out of a myriad of different molecules.[5] Jacques Monod attributed a teleonomic performance or function to these biological complexes. Teleonomy implies the idea of an oriented, coherent and constructive activity. Proteins therefore must be considered essential molecular agents in the teleonomic performances of all living beings.[6]
Affinity
The highest possible affinity from a protein towards the ligand, or target molecule, can be observed when the protein has a perfect mirror image of the shape of the target surface together with a charge distribution that complements perfectly the target surface.[7] The affinity between protein and ligand is given by the equilibrium dissociation constant Kd or the inverse of the association constant 1/Ka (or binding constant 1/Kb) that relates the concentrations of the complexed and uncomplexed species in solution.
The dissociation constant is defined as
Kd =
where [L], [P] and [LP] represent molar concentrations of the protein, ligand and complex, respectively.
The lower the Kd value the higher the affinity of the protein for the ligand and vice versa. The Kd value is equivalent to the concentration of the ligand at which one-half of the proteins contain bound ligand.[3][8] Affinity is influenced also by the properties of the solution, like pH, temperature and salt concentration, that may affect the stable state of the proteins and ligands and hence also their interaction and by the presence of other macromolecules that causes macromolecular crowding.[9]
Functions
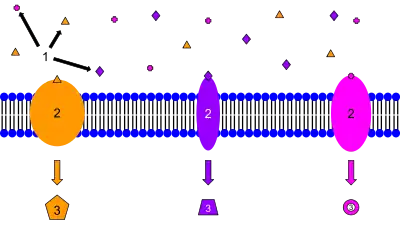
Protein–ligand complexes can be found in almost any cellular process. Binding of a ligand causes a conformational change in the protein and often also in the ligand. This change initiates a sequence of events leading to different cellular functions. The complexes are formed by different molecules like macromolecules as in protein complexes, protein DNA or protein RNA complexes as well as by proteins that bind smaller molecules like peptides, lipids, carbohydrates, small nucleic acids. They may have various functions within the cell: catalysis of chemical reactions (enzyme-substrate), defense of the organism through the immune system (antibodies antigen complexes), signal transduction (receptor-ligand complexes) that consists of a transmembrane receptor that upon binding the ligand activates an intracellular cascade. Lipophilic hormonal receptor complexes can pass the nuclear membrane where transcription may be regulated.[8]
Example
Protein-Ligand complex is essential in many of the cellular processes that occur within organisms. One of these examples is the Glucagon receptor (GCGR). Glucagon receptor (GCGR) is a family of G-protein coupled receptors (GPCRs) in humans that plays an important role in maintaining glucose concentration within the blood during periods of low energy state. Glucagon binding to GPCRcauses a conformational change in the intracellular domain, allowing interaction with the heterotrimeric Gs protein. The alpha Subunit of the Gs protein releases bound GDP and binds GTP. The alpha subunit-GTP complex dissociates from the beta and gamma dimer and interacts with adenylate cyclase. Binding of glucagon molecule activates many of the alpha subunit, which amplifies the hormonal signal. Then, the alpha subunit activates the adenylate cyclase, which converts ATP to cAMP. The alpha subunit deactivates itself within minutes by hydrolyzing GTP to GDP (GTPase activity). The alpha subunit reassociates with beta-gamma dimer to form an inactive complex. A better understanding of the protein-ligand complex mechanisms may allow us for the treatment of some diseases such as type 2 diabetes.[10] Glucagon receptor inhibitors are promising for the treatment of type 2 diabetes.[11] Inhibitors of Glucagon receptors are either glucagon neutralizers or small molecular antagonists, and they all rely on the concept of protein-ligand complex interaction.[11]
See also
- Dissociation constant
- Ligand (biochemistry)
- Receptor (biochemistry)
References
- ↑ Bohl CE, Gao W, Miller DD, Bell CE, Dalton JT (April 2005). "Structural basis for antagonism and resistance of bicalutamide in prostate cancer". Proceedings of the National Academy of Sciences of the United States of America. 102 (17): 6201–6. Bibcode:2005PNAS..102.6201B. doi:10.1073/pnas.0500381102. PMC 1087923. PMID 15833816.
- ↑ Fragment Based Drug Design: Tools, Practical Approaches, and Examples. Academic Press. 28 February 2011. pp. 265–. ISBN 978-0-12-381275-9.
- 1 2 Du X, Li Y, Xia YL, Ai SM, Liang J, Sang P, Ji XL, Liu SQ (January 2016). "Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods". International Journal of Molecular Sciences. 17 (2): 144. doi:10.3390/ijms17020144. PMC 4783878. PMID 26821017.
- ↑ Bongrand P (1999). "Ligand-receptor interactions". Reports on Progress in Physics. 62 (6): 921–968. arXiv:0809.1926. Bibcode:1999RPPh...62..921B. doi:10.1088/0034-4885/62/6/202. S2CID 41417093.
- ↑ R., Loewenstein, Werner (2013-01-29). Physics in mind : a quantum view of the brain. New York. ISBN 978-0-465-02984-6. OCLC 778420640.
- ↑ Monod J (1970). Le hasard et la nécessité. Essai sur la philosophie naturelle de la biologie moderne [Chance and necessity Essay on the natural philosophy of modern biology] (in French). Le Seuil.
- ↑ Eaton BE, Gold L, Zichi DA (October 1995). "Let's get specific: the relationship between specificity and affinity". Chemistry & Biology. 2 (10): 633–8. doi:10.1016/1074-5521(95)90023-3. PMID 9383468.
- 1 2 Lodish H (1996). Molecular Cell Biology. Scientific American Books. pp. 854–918.
- ↑ Zhou HX, Rivas G, Minton AP (2008). "Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences". Annual Review of Biophysics. 37: 375–97. doi:10.1146/annurev.biophys.37.032807.125817. PMC 2826134. PMID 18573087.
- ↑ Janah, Lina; Kjeldsen, Sasha; Galsgaard, Katrine D.; Winther-Sørensen, Marie; Stojanovska, Elena; Pedersen, Jens; Knop, Filip K.; Holst, Jens J.; Wewer Albrechtsen, Nicolai J. (2019-07-05). "Glucagon Receptor Signaling and Glucagon Resistance". International Journal of Molecular Sciences. 20 (13): 3314. doi:10.3390/ijms20133314. ISSN 1422-0067. PMC 6651628. PMID 31284506.
- 1 2 Baig, M. H.; Ahmad, K.; Hasan, Q.; Khan, M. K. A.; Rao, N. S.; Kamal, M. A.; Choi, I. (2015). "Interaction of Glucagon G-Protein Coupled Receptor with Known Natural Antidiabetic Compounds: Multiscoring In Silico Approach". Evidence-Based Complementary and Alternative Medicine. 2015: 497253. doi:10.1155/2015/497253. ISSN 1741-427X. PMC 4508340. PMID 26236379.