Thermal cycler

The thermal cycler (PCR machine or DNA amplifier) is a laboratory apparatus most commonly used to amplify segments of DNA via the polymerase chain reaction (PCR).[1]
Thermal cyclers may also be used in laboratories to facilitate other temperature-sensitive reactions, including restriction enzyme digestion or rapid diagnostics.[2]
The device has a thermal block with holes where tubes holding the reaction mixtures can be inserted. The cycler then raises and lowers the temperature of the block in discrete, pre-programmed steps.[3][4]
History
Initial thermal cyclers were designed for use with the Klenow fragment of DNA polymerase I. Since this enzyme is destroyed during each heating step of the amplification process, new enzyme had to be added every cycle. This led to a cumbersome machine based on an automated pipettor, with open reaction tubes. Later, the PCR process was adapted to the use of thermostable DNA polymerase from Thermus aquaticus, which greatly simplified the design of the thermal cycler.[5][6][7]
In modern PCR machines a Peltier element is commonly used; quality thermal cyclers often contain silver blocks to achieve fast temperature changes and uniform temperature throughout the block. Other cyclers have multiple blocks with high heat capacity, each of which is kept at a constant temperature, and the reaction tubes are moved between them by means of an automated process. Miniaturized thermal cyclers have been created in which the reaction mixture moves via channel through hot and cold zones on a microfluidic chip. Thermal cyclers designed for quantitative PCR have optical systems which enable fluorescence to be monitored during reaction cycling.[8][9][10]
Modern innovations
 Biometra TRIO, a thermal cycler with touchscreen interface
Biometra TRIO, a thermal cycler with touchscreen interface PCR tubes
PCR tubes A thermal cycler with lid opened
A thermal cycler with lid opened
Modern thermal cyclers are equipped with a heated lid that presses against the lids of the reaction tubes. This prevents condensation of water from the reaction mixtures on the insides of the lids. Traditionally, a layer of mineral oil was used for this purpose. Some thermal cyclers are equipped with a fully adjustable heated lid to allow for nonstandard or diverse types of PCR plasticware.[11]
Some thermal cyclers are equipped with multiple blocks allowing several different PCRs to be carried out simultaneously. Some models also have a gradient function to allow for different temperatures in different parts of the block. [12]
Medical and diagnostic applications
Prospective parents can be tested for being genetic carriers, or their children might be tested for actually being affected by a disease.[13] DNA samples for prenatal testing can be obtained by amniocentesis, chorionic villus sampling, or even by the analysis of rare fetal cells circulating in the mother's bloodstream.[14][15] PCR analysis is also essential to preimplantation genetic diagnosis, where individual cells of a developing embryo are tested for mutations.[16]
- PCR can also be used as part of a sensitive test for tissue typing, vital to organ transplantation. As of 2008, there is even a proposal to replace the traditional antibody-based tests for blood type with PCR-based tests.[17]
- Many forms of cancer involve alterations to oncogenes. By using PCR-based tests to study these mutations, therapy regimens can sometimes be individually customized to a patient. PCR permits early diagnosis of malignant diseases such as leukemia and lymphomas, which is currently the highest-developed in cancer research and is already being used routinely. PCR assays can be performed directly on genomic DNA samples to detect translocation-specific malignant cells at a sensitivity that is at least 10,000 fold higher than that of other methods.[18] PCR is very useful in the medical field since it allows for the isolation and amplification of tumor suppressors. Quantitative PCR for example, can be used to quantify and analyze single cells, as well as recognize DNA, mRNA and protein confirmations and combinations.[19]
Infectious disease applications
PCR allows for rapid and highly specific diagnosis of infectious diseases, including those caused by bacteria or viruses.[20] PCR also permits identification of non-cultivatable or slow-growing microorganisms such as mycobacteria, anaerobic bacteria, or viruses from tissue culture assays and animal models. The basis for PCR diagnostic applications in microbiology is the detection of infectious agents and the discrimination of non-pathogenic from pathogenic strains by virtue of specific genes.[20][21]
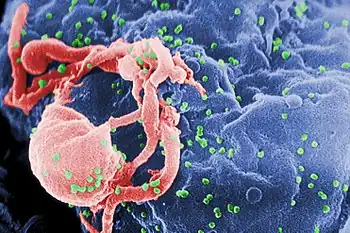
Characterization and detection of infectious disease organisms have been revolutionized by PCR in the following ways:
- The human immunodeficiency virus (or HIV), is a difficult target to find and eradicate. The earliest tests for infection relied on the presence of antibodies to the virus circulating in the bloodstream. However, antibodies don't appear until many weeks after infection, maternal antibodies mask the infection of a newborn, and therapeutic agents to fight the infection don't affect the antibodies. PCR tests have been developed that can detect as little as one viral genome among the DNA of over 50,000 host cells.[22]
- The spread of a disease organism through populations of domestic or wild animals can be monitored by PCR testing. In many cases, the appearance of new virulent sub-types can be detected and monitored.[23]
- Viral DNA can be detected by PCR. The primers used must be specific to the targeted sequences in the DNA of a virus, and PCR can be used for diagnostic analyses or DNA sequencing of the viral genome. The high sensitivity of PCR permits virus detection soon after infection and even before the onset of disease.[20] Such early detection may give physicians a significant lead time in treatment. The amount of virus ("viral load") in a patient can also be quantified by PCR-based DNA quantitation techniques (see below). A variant of PCR (RT-PCR) is used for detecting viral RNA rather than DNA: in this test the enzyme reverse transcriptase is used to generate a DNA sequence which matches the viral RNA; this DNA is then amplified as per the usual PCR method. RT-PCR is widely used to detect the SARS-CoV-2 viral genome.[24]
- Diseases such as pertussis (or whooping cough) are caused by the bacteria Bordetella pertussis. This bacteria is marked by a serious acute respiratory infection that affects various animals and humans and has led to the deaths of many young children. The pertussis toxin is a protein exotoxin that binds to cell receptors by two dimers and reacts with different cell types such as T lymphocytes which play a role in cell immunity.[25] PCR is an important testing tool that can detect sequences within the gene for the pertussis toxin. Because PCR has a high sensitivity for the toxin and a rapid turnaround time, it is very efficient for diagnosing pertussis when compared to culture.[26]
References
- ↑ Weier, HU; Gray, JW (Jul–Aug 1988). "A programmable system to perform the polymerase chain reaction". DNA (Mary Ann Liebert, Inc.). 7 (6): 441–7. PMID 3203600.
- ↑ Higgins, James A.; Nasarabadi, Shanavaz; Karns, Jeffrey S.; Shelton, Daniel R.; Cooper, Mary; Gbakima, Aiah; Koopman, Ronald P. (1 August 2003). "A handheld real time thermal cycler for bacterial pathogen detection". Biosensors and Bioelectronics. 18 (9): 1115–1123. doi:10.1016/S0956-5663(02)00252-X.
- ↑ "Thermal Cycler (PCR Machine)". www.iitk.ac.in. Archived from the original on 27 September 2022. Retrieved 24 September 2022.
- ↑ Pelt-Verkuil, Elizabeth van; Belkum, Alex van; Hays, John P. (14 March 2008). Principles and Technical Aspects of PCR Amplification. Springer Science & Business Media. p. 20. ISBN 978-1-4020-6241-4. Archived from the original on 27 September 2022. Retrieved 26 September 2022.
- ↑ "The Original Micropipette". The Scientist Magazine®. Archived from the original on 25 September 2021. Retrieved 25 September 2022.
- ↑ Pelt-Verkuil, Elizabeth van; Belkum, Alex van; Hays, John P. (14 March 2008). Principles and Technical Aspects of PCR Amplification. Springer Science & Business Media. p. 103. ISBN 978-1-4020-6241-4. Retrieved 28 September 2022.
- ↑ Briggs, George M. (2021). "Thermus aquaticus". milnepublishing. Archived from the original on 18 April 2022. Retrieved 3 October 2022.
- ↑ "Peltiers in PCR matter! - Eppendorf Handling Solutions". handling-solutions.eppendorf.com. Archived from the original on 27 September 2022. Retrieved 26 September 2022.
- ↑ Kubista, Mikael; Andrade, José Manuel; Bengtsson, Martin; Forootan, Amin; Jonák, Jiri; Lind, Kristina; Sindelka, Radek; Sjöback, Robert; Sjögreen, Björn; Strömbom, Linda; Ståhlberg, Anders; Zoric, Neven (April 2006). "The real-time polymerase chain reaction". Molecular Aspects of Medicine. 27 (2–3): 95–125. doi:10.1016/j.mam.2005.12.007. ISSN 0098-2997. Archived from the original on 24 May 2022. Retrieved 29 September 2022.
- ↑ Zou, Quanbo; Sridhar, Uppili; Chen, Yu; Lim, Tit Meng; Zachariah, Emmanuel Selvanayagam; Yan, Tie (18 February 2003). "Miniaturized thermal cycler". Archived from the original on 6 May 2021. Retrieved 2 October 2022.
- ↑ "The Most "Advanced" Thermal Cycler Yet - Get Informed". Get Informed. 2014-09-29. Archived from the original on 2018-01-31. Retrieved 2017-05-31.
- ↑ Rupprecht, Charles C. E.; Nagarajan, Thirumeni (9 February 2015). Current Laboratory Techniques in Rabies Diagnosis, Research and Prevention, Volume 2. Academic Press. p. 122. ISBN 978-0-12-802012-8. Archived from the original on 29 September 2022. Retrieved 29 September 2022.
- ↑ Saiki, Randall K.; Scharf, Stephen; Faloona, Fred; Mullis, Kary B.; Horn, Glenn T.; Erlich, Henry A.; Arnheim, Norman (20 December 1985). "Enzymatic Amplification of β-Globin Genomic Sequences and Restriction Site Analysis for Diagnosis of Sickle Cell Anemia". Science. 230 (4732): 1350–1354. doi:10.1126/science.2999980. ISSN 0036-8075. Archived from the original on 18 March 2022. Retrieved 23 September 2022.
- ↑ Nicolini, Umberto; Lalatta, Faustina; Natacci, Federica; Curcio, Cristina; Bui, The-Hung (November 2004). "The introduction of QF-PCR in prenatal diagnosis of fetal aneuploidies: time for reconsideration". Human Reproduction Update. 10 (6): 541–548. doi:10.1093/humupd/dmh046. ISSN 1355-4786. Archived from the original on 2022-03-27. Retrieved 2022-09-24.
- ↑ "Prenatal Genetic Diagnostic Tests". www.acog.org. Archived from the original on 28 July 2022. Retrieved 27 September 2022.
- ↑ Dreesen, Jos; Destouni, Aspasia; Kourlaba, Georgia; Degn, Birte; Mette, Wulf Christensen; Carvalho, Filipa; Moutou, Celine; Sengupta, Sioban; Dhanjal, Seema; Renwick, Pamela; Davies, Steven; Kanavakis, Emmanouel; Harton, Gary; Traeger-Synodinos, Joanne (August 2014). "Evaluation of PCR-based preimplantation genetic diagnosis applied to monogenic diseases: a collaborative ESHRE PGD consortium study". European journal of human genetics: EJHG. 22 (8): 1012–1018. doi:10.1038/ejhg.2013.277. ISSN 1476-5438. Archived from the original on 18 June 2022. Retrieved 25 September 2022.
- ↑ Quill E (March 2008). "Medicine. Blood-matching goes genetic". Science. 319 (5869): 1478–79. doi:10.1126/science.319.5869.1478. PMID 18339916. S2CID 36945291.
- ↑ Tomar, Rukam (2010). Molecular Markers and Plant Biotechnology. Pitman Pura, New Delhi: New India Publishing Agency. p. 188. ISBN 978-93-80235-25-7.
- ↑ Garibyan, Lilit; Avashia, Nidhi (March 2013). "Polymerase chain reaction". The Journal of Investigative Dermatology. 133 (3): 1–4. doi:10.1038/jid.2013.1. ISSN 1523-1747. Archived from the original on 15 June 2022. Retrieved 23 September 2022.
- 1 2 3 Cai HY, Caswell JL, Prescott JF (March 2014). "Nonculture molecular techniques for diagnosis of bacterial disease in animals: a diagnostic laboratory perspective". Veterinary Pathology. 51 (2): 341–50. doi:10.1177/0300985813511132. PMID 24569613.
- ↑ Salis AD (2009). "Applications in Clinical Microbiology". Real-Time PCR: Current Technology and Applications. Caister Academic Press. ISBN 978-1-904455-39-4.
- ↑ Kwok S, Mack DH, Mullis KB, Poiesz B, Ehrlich G, Blair D, et al. (May 1987). "Identification of human immunodeficiency virus sequences by using in vitro enzymatic amplification and oligomer cleavage detection". Journal of Virology. 61 (5): 1690–94. doi:10.1128/jvi.61.5.1690-1694.1987. PMC 254157. PMID 2437321.
- ↑ M.p, Ward; S.w, Laffan; L.d, Highfield (August 2011). "Disease spread models in wild and feral animal populations: application of artificial life models". 30 (2): 437. doi:10.20506/rst.30.2.2042. ISSN 0253-1933. Archived from the original on 28 September 2022. Retrieved 28 September 2022.
{{cite journal}}: Cite journal requires|journal=(help) - ↑ "Coronavirus: il viaggio dei test". Istituto Superiore di Sanità. Archived from the original on 2022-06-12. Retrieved 2022-09-23.
- ↑ Finger, Horst; von Koenig, Carl Heinz Wirsing (1996). Baron, Samuel (ed.). Medical Microbiology (4th ed.). Galveston, TX: University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. PMID 21413270. Archived from the original on 2019-05-03. Retrieved 2022-09-23.
- ↑ Yeh, Sylvia H.; Mink, ChrisAnna M. (2012). "Bordetella pertussis and Pertussis (Whooping Cough)". Netter's Infectious Diseases. Netter's Infectious Diseases. pp. 11–14. doi:10.1016/B978-1-4377-0126-5.00003-3. ISBN 978-1-4377-0126-5.