Viral replication
| Influenza virus life cycle |
|---|
 |
|
Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.[1]
Viral production / replication
Viruses multiply only in living cells. The host cell must provide the energy and synthetic machinery and the low- molecular-weight precursors for the synthesis of viral proteins and nucleic acids.[2]
The virus replication occurs in seven stages, namely;
- Attachment
- Entry,
- Uncoating,
- Transcription / mRNA production,
- Synthesis of virus components,
- Virion assembly and
- Release (Liberation Stage).
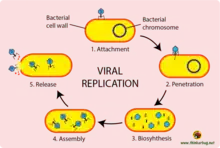 Viral replication of a bacteriophage[3]
Viral replication of a bacteriophage[3]
Attachment
It is the first step of viral replication. The virus attaches to the cell membrane of the host cell. It then injects its DNA or RNA into the host to initiate infection. In animal cells these viruses get into the cell through the process of endocytosis which works through fusing of the virus and fusing of the viral envelope with the cell membrane of the animal cell and in plant cells it enters through the process of pinocytosis which works on pinching of the viruses.
Entry
The cell membrane of the host cell invaginates the virus particle, enclosing it in a pinocytotic vacuole. This protects the cell from antibodies like in the case of the HIV virus.
Uncoating
Cell enzymes (from lysosomes) strip off the virus protein coat. This releases or renders accessible the virus nucleic acid or genome.
Transcription / mRNA production
For some RNA viruses, the infecting RNA produces messenger RNA (mRNA), which can translate the genome into protein products. For viruses with negative stranded RNA, or DNA, viruses are produced by transcription then translation.
The mRNA is used to instruct the host cell to make virus components. The virus takes advantage of the existing cell structures to replicate itself.
Synthesis of virus components
The components are manufactured by the virus using the host's existing organelles:
- Viral proteins: Viral mRNA is translated on cellular ribosomes into two types of viral protein:
- Structural: proteins which make up the virus particle
- Nonstructural: proteins not found in the virus particle, mainly enzymes for virus genome replication
- Viral nucleic acid (genome replication): New viral genomes are synthesized; templates are either the parental genome or newly formed complementary strands, in the case of single-stranded genomes. These genomes are made by either a viral polymerase or (in some DNA viruses) a cellular enzyme, particularly in rapidly dividing cells.
Virion assembly
A virion is simply an active or intact virus particle. In this stage, newly synthesized genome (nucleic acid), and proteins are assembled to form new virus particles.
This may take place in the cell's nucleus, cytoplasm, or at plasma membrane for most developed viruses.
Release (liberation stage)
The viruses, now being matured are released by either sudden rupture of the cell, or gradual extrusion (force out) of enveloped viruses through the cell membrane.
The new viruses may invade or attack other cells, or remain dormant in the cell. In the case of bacterial viruses, the release of progeny virions takes place by lysis of the infected bacterium. However, in the case of animal viruses, release usually occurs without cell lysis.
Baltimore Classification System
Viruses are classed into 7 types of genes, each of which has its own families of viruses, which in turn have differing replication strategies themselves.[4] David Baltimore, a Nobel Prize-winning biologist, devised a system called the Baltimore Classification System to classify different viruses based on their unique replication strategy. There are seven different replication strategies based on this system (Baltimore Class I, II, III, IV, V, VI, VII). The seven classes of viruses are listed here briefly and in generalities.[5]
Class 1: Double-stranded DNA viruses
This type of virus usually must enter the host nucleus before it is able to replicate. Some of these viruses require host cell polymerases to replicate their genome, while others, such as adenoviruses or herpes viruses, encode their own replication factors. However, in either cases, replication of the viral genome is highly dependent on a cellular state permissive to DNA replication and, thus, on the cell cycle. The virus may induce the cell to forcefully undergo cell division, which may lead to transformation of the cell and, ultimately, cancer. An example of a family within this classification is the Adenoviridae.
There is only one well-studied example in which a class 1 family of viruses does not replicate within the nucleus. This is the Poxvirus family, which comprises highly pathogenic viruses that infect vertebrates.
Class 2: Single-stranded DNA viruses
Viruses that fall under this category include ones that are not as well-studied, but still do pertain highly to vertebrates. Two examples include the Circoviridae and Parvoviridae. They replicate within the nucleus, and form a double-stranded DNA intermediate during replication. A human Anellovirus called TTV is included within this classification and is found in almost all humans, infecting them asymptomatically in nearly every major organ.
Class 3: Double-stranded RNA viruses
Like most viruses with RNA genomes, double-stranded RNA viruses do not rely on host polymerases for replication to the extent that viruses with DNA genomes do. Double-stranded RNA viruses are not as well-studied as other classes. This class includes two major families, the Reoviridae and Birnaviridae. Replication is monocistronic and includes individual, segmented genomes, meaning that each of the genes codes for only one protein, unlike other viruses, which exhibit more complex translation.
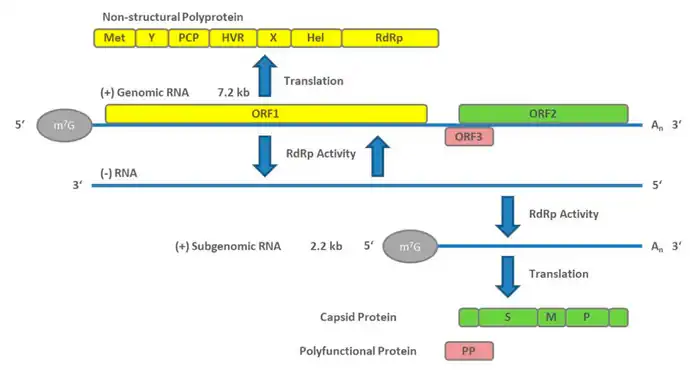
Classes 4 & 5: Single-stranded RNA viruses
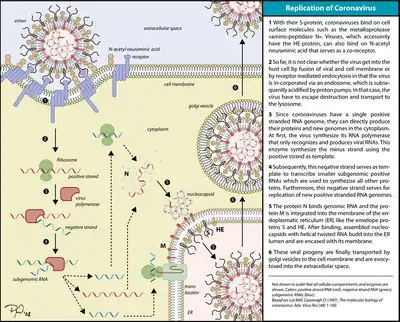
These viruses consist of two types, however both share the fact that replication is primarily in the cytoplasm, and that replication is not as dependent on the cell cycle as that of DNA viruses. This class of viruses is also one of the most-studied types of viruses, alongside the double-stranded DNA viruses.
Class 4: Single-stranded RNA viruses - positive-sense
The positive-sense RNA viruses and indeed all genes defined as positive-sense can be directly accessed by host ribosomes to immediately form proteins. These can be divided into two groups, both of which replicate in the cytoplasm:
- Viruses with polycistronic mRNA where the genome RNA forms the mRNA and is translated into a polyprotein product that is subsequently cleaved to form the mature proteins. This means that the gene can utilize a few methods in which to produce proteins from the same strand of RNA, reducing the size of its genome.
- Viruses with complex transcription, for which subgenomic mRNAs, ribosomal frameshifting and proteolytic processing of polyproteins may be used. All of which are different mechanisms with which to produce proteins from the same strand of RNA.
Examples of this class include the families Coronaviridae, Flaviviridae, and Picornaviridae.
Class 5: Single-stranded RNA viruses - negative-sense
The negative-sense RNA viruses and indeed all genes defined as negative-sense cannot be directly accessed by host ribosomes to immediately form proteins. Instead, they must be transcribed by viral polymerases into the "readable" complementary positive-sense. These can also be divided into two groups:
- Viruses containing nonsegmented genomes for which the first step in replication is transcription from the negative-stranded genome by the viral RNA-dependent RNA polymerase to yield monocistronic mRNAs that code for the various viral proteins. A positive-sense genome copy that serves as template for production of the negative-strand genome is then produced. Replication is within the cytoplasm.
- Viruses with segmented genomes for which replication occurs in the cytoplasm and for which the viral RNA-dependent RNA polymerase produces monocistronic mRNAs from each genome segment.
Examples in this class include the families Orthomyxoviridae, Paramyxoviridae, Bunyaviridae, Filoviridae, and Rhabdoviridae (which includes rabies).
Class 6: Positive-sense single-stranded RNA viruses that replicate through a DNA intermediate
A well-studied family of this class of viruses include the retroviruses. One defining feature is the use of reverse transcriptase to convert the positive-sense RNA into DNA. Instead of using the RNA for templates of proteins, they use DNA to create the templates, which is spliced into the host genome using integrase. Replication can then commence with the help of the host cell's polymerases.
Class 7: Double-stranded DNA viruses that replicate through a single-stranded RNA intermediate
This small group of viruses, exemplified by the Hepatitis B virus, have a double-stranded, gapped genome that is subsequently filled in to form a covalently closed circle (cccDNA) that serves as a template for production of viral mRNAs and a subgenomic RNA. The pregenome RNA serves as template for the viral reverse transcriptase and for production of the DNA genome.
References
- ↑ Roberts RJ, "Fish pathology, 3rd Edition", Elsevier Health Sciences, 2001.
- ↑ Geo.F. Brooks, M.D et al. "Jawetz, Melnick & Adelberg's MEDICAL MICROBIOLOGY.pdf, 26th Edition, McGraw Hill, 2013, ISBN 978-0-07-181578-9
- ↑ "Viral replication". THINKER BUG. Retrieved 2021-09-11.
- ↑ Girdhar, Khyati; Powis, Amaya; Raisingani, Amol; Chrudinová, Martina; Huang, Ruixu; Tran, Tu; Sevgi, Kaan; Dogus Dogru, Yusuf; Altindis, Emrah (29 September 2021). "Viruses and Metabolism: The Effects of Viral Infections and Viral Insulins on Host Metabolism". Annual Review of Virology. 8 (1): 373–391. doi:10.1146/annurev-virology-091919-102416. ISSN 2327-056X. PMC 9175272. PMID 34586876.
- ↑ N.J. Dimmock et al. "Introduction to Modern Virology, 6th edition." Blackwell Publishing, 2007.
