FCAR
Fc fragment of IgA receptor (FCAR) is a human gene[3] that codes for the transmembrane receptor FcαRI, also known as CD89 (Cluster of Differentiation 89). FcαRI binds the heavy-chain constant region of Immunoglobulin A (IgA) antibodies.[4] FcαRI is present on the cell surface of myeloid lineage cells, including neutrophils, monocytes, macrophages, and eosinophils,[5] though it is notably absent from intestinal macrophages[6] and does not appear on mast cells.[5] FcαRI plays a role in both pro- and anti-inflammatory responses depending on the state of IgA bound.[5] Inside-out signaling primes FcαRI in order for it to bind its ligand,[4] while outside-in signaling caused by ligand binding depends on FcαRI association with the Fc receptor gamma chain (FcR γ-chain).[5]
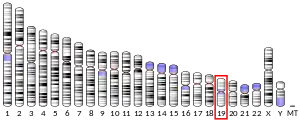
Though FcαRI is part of the Fc receptor immunoglobulin superfamily, the protein's primary structure is similar to receptors in the leukocyte receptor cluster (LRC), and the FCAR gene appears amidst LRC genes on chromosome 19.[4][5] This contrasts with the location of other members of the Fc receptor immunoglobulin superfamily, which are encoded on chromosome 1.[4][5] Additionally, though there are equivalents to FCAR in several species, there is no such homolog in mice.[4]
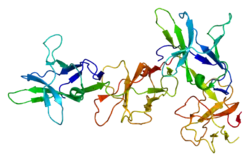
Structure
The FcαRI α-chain consists of two extracellular domains, EC1 and EC2, at a right angle to each other, a transmembrane domain, and an intracellular domain.[4] However, this chain alone cannot perform signaling in response to IgA binding, and FcαRI must associate with a dimeric form of FcR g-chain, the ends of which contain immunoreceptor tyrosine-based activation motifs (ITAMs). The FcR γ-chain is responsible for relaying the signal to the inside of the cell.[4][5]
Two FCAR alleles differing by a single nucleotide polymorphism (SNP) code for two FcαRI molecules that differ in their ability to signal for IL-6 and TNF-α production and release.[7] The SNP results in either serine or glycine as the 248th residue of the amino acid sequence, a position in the intracellular domain of FcαRI.[7] Compared to FcαRI with Ser248, FcαRI molecules with Gly248 are better able to signal for the release of IL-6, even independently from FcR γ-chain association.[7]
Alternative splicing of the transcript from this gene produces ten mRNA variants encoding different isoforms.[3]
Inside-Out Signaling
FcαRI must first be primed by a process called inside-out signaling in order to bind with increased ability to IgA. Priming occurs when cytokines signaling the presence of an infection bind their receptors on FcαRI-expressing cells, activating the kinase PI3K. PI3K then activates p38 and PKC, which together with PP2A lead to the dephosphorylation of the Serine 263 residue (Ser263) on the intracellular domain of the FcαRI α-chain.[8] The priming of FcαRI to be able to bind IgA does not depend on FcαRI association with the FcR γ-chain,[5] but does depend on cytoskeleton organization.[8]
Once primed, FcαRI can bind IgA.[8] The FcαRI EC1 domain binds the hinge between the IgA-Fc regions Ca2 and Ca3 regions.[4]
Function
Signaling and the resulting cellular response caused by FcαRI binding IgA varies depending on the state of the IgA molecules. A pro-inflammatory response is signaled when IgA molecules in an immune complex bind to multiple FcαRI, resulting in the activation of Src family kinases and the phosphorylation of the FcR γ-chain ITAMs by Lyn.[9] Syk, a tyrosine kinase, subsequently docks at the phosphorylated ITAMs and initiates PI3K and PLC-γ signaling.[9] The ensuing signaling cascades lead to pro-inflammatory responses such as release of cytokines, phagocytosis, respiratory bursts, antibody-dependent cell-mediated cytotoxicity, production of reactive oxygen species, and antigen presentation.[4][5]
Despite signaling via ITAMs, which typically initiate activation cascades, FcαRI may either act as an activating or inhibitory receptor.[10] Inhibitory ITAM signaling (ITAMi) results in anti-inflammatory responses. When FcαRI monovalently binds monomeric, non-antigen bound IgA, the form most common in serum,[4] the resulting signals result in inactivation of other activating receptors such as FcγR and FcεRI. The binding of the monomeric serum IgA causes Lyn to only partly phosphorylate the FcR γ-chain ITAMs. Consequently, Src homology region 2 domain-containing phosphatase-1 (SHP-1) is recruited by Syk to the FcR γ-chain.[9] A tyrosine phosphatase, SHP-1 coordinates the anti-inflammatory response, preventing other receptors from signaling for pro-inflammatory responses by not allowing these receptors to become phosphorylated.[9] This ITAMi signaling supports homeostasis in the absence of pathogens.[9]
The anti-inflammatory role of monomeric IgA-FcαRI binding may have implications for treatment of allergic asthma, as shown by targeting FcαRI in transgenic mice models with anti-FcαRI Fab antibodies, which mimic the binding of monomeric IgA.[11] This FcαRI targeting led to decreased infiltration of airway tissue by inflammatory leukocytes.[11]
The secreted form of IgA (sIgA), a homodimer secreted across epithelial linings such as the gut epithelium, is sterically hindered in its binding to FcαRI. This is because some of sIgA's FcαRI binding site is obscured by a section of the cleaved polymeric Ig receptor that aided sIgA's secretion into the gut lumen.[5] However, the precursor to sIgA, dimeric IgA (dIgA), binds to FcαRI with approximately the same affinity as monomeric IgA.[5] Secreted IgA plays an important role in preventing immune response to commensal gut microbes, and accordingly intestinal macrophages do not express FcαRI.[4] However, during invasion of mucosal tissue by pathogenic bacteria, neutrophils responding to the infection will bind and phagocytose dIgA-opsonized bacteria via FcαRI.[4]
FcαRI is also an important Fc receptor for neutrophil killing of tumor cells. When FcαRI-expressing neutrophils come into contact with IgA-opsonized tumor cells, the neutrophils not only perform antibody-dependent cell-mediated cytotoxicity, but also release the cytokines TNF-α and IL-1β which cause increased neutrophil migration to the site.[12]
See also
References
- ENSG00000284004, ENSG00000273738, ENSG00000283953, ENSG00000276985, ENSG00000275970, ENSG00000284061, ENSG00000283750, ENSG00000278415, ENSG00000186431, ENSG00000275136, ENSG00000276858, ENSG00000284245, ENSG00000275269, ENSG00000274580 GRCh38: Ensembl release 89: ENSG00000275564, ENSG00000284004, ENSG00000273738, ENSG00000283953, ENSG00000276985, ENSG00000275970, ENSG00000284061, ENSG00000283750, ENSG00000278415, ENSG00000186431, ENSG00000275136, ENSG00000276858, ENSG00000284245, ENSG00000275269, ENSG00000274580 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: FCAR Fc fragment of IgA, receptor for".
- Bakema JE, van Egmond M (November 2011). "The human immunoglobulin A Fc receptor FcαRI: a multifaceted regulator of mucosal immunity". Mucosal Immunology. 4 (6): 612–24. doi:10.1038/mi.2011.36. PMID 21937986.
- Aleyd E, Heineke MH, van Egmond M (November 2015). "The era of the immunoglobulin A Fc receptor FcαRI; its function and potential as target in disease". Immunological Reviews. 268 (1): 123–38. doi:10.1111/imr.12337. PMID 26497517. S2CID 21300390.
- Smith PD, Smythies LE, Mosteller-Barnum M, Sibley DA, Russell MW, Merger M, Sellers MT, Orenstein JM, Shimada T, Graham MF, Kubagawa H (September 2001). "Intestinal macrophages lack CD14 and CD89 and consequently are down-regulated for LPS- and IgA-mediated activities". Journal of Immunology. 167 (5): 2651–6. doi:10.4049/jimmunol.167.5.2651. PMID 11509607.
- Wu J, Ji C, Xie F, Langefeld CD, Qian K, Gibson AW, Edberg JC, Kimberly RP (March 2007). "FcalphaRI (CD89) alleles determine the proinflammatory potential of serum IgA". Journal of Immunology. 178 (6): 3973–82. doi:10.4049/jimmunol.178.6.3973. PMID 17339498.
- Brandsma AM, Jacobino SR, Meyer S, ten Broeke T, Leusen JH (November 2015). "Fc receptor inside-out signaling and possible impact on antibody therapy". Immunological Reviews. 268 (1): 74–87. doi:10.1111/imr.12332. PMID 26497514. S2CID 20610179.
- Mkaddem SB, Rossato E, Heming N, Monteiro RC (April 2013). "Anti-inflammatory role of the IgA Fc receptor (CD89): from autoimmunity to therapeutic perspectives". Autoimmunity Reviews. 12 (6): 666–9. doi:10.1016/j.autrev.2012.10.011. PMID 23201915.
- Getahun A, Cambier JC (November 2015). "Of ITIMs, ITAMs, and ITAMis: revisiting immunoglobulin Fc receptor signaling". Immunological Reviews. 268 (1): 66–73. doi:10.1111/imr.12336. PMC 4621791. PMID 26497513.
- Pasquier B, Launay P, Kanamaru Y, Moura IC, Pfirsch S, Ruffié C, Hénin D, Benhamou M, Pretolani M, Blank U, Monteiro RC (January 2005). "Identification of FcalphaRI as an inhibitory receptor that controls inflammation: dual role of FcRgamma ITAM". Immunity. 22 (1): 31–42. doi:10.1016/j.immuni.2004.11.017. PMID 15664157.
- van Egmond M, Bakema JE (June 2013). "Neutrophils as effector cells for antibody-based immunotherapy of cancer". Seminars in Cancer Biology. 23 (3): 190–9. doi:10.1016/j.semcancer.2012.12.002. PMID 23287459.
- Morton HC, van den Herik-Oudijk IE, Vossebeld P, Snijders A, Verhoeven AJ, Capel PJ, van de Winkel JG (December 1995). "Functional association between the human myeloid immunoglobulin A Fc receptor (CD89) and FcR gamma chain. Molecular basis for CD89/FcR gamma chain association". The Journal of Biological Chemistry. 270 (50): 29781–7. doi:10.1074/jbc.270.50.29781. PMID 8530370.
Further reading
- Morton HC, van Egmond M, van de Winkel JG (1996). "Structure and function of human IgA Fc receptors (Fc alpha R)". Critical Reviews in Immunology. 16 (4): 423–40. PMID 8954257.
- Morton HC, Brandtzaeg P (2001). "CD89: the human myeloid IgA Fc receptor". Archivum Immunologiae et Therapiae Experimentalis. 49 (3): 217–29. PMID 11478396.
- Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT (February 2002). "Leukocyte Ig-like receptor complex (LRC) in mice and men". Trends in Immunology. 23 (2): 81–8. doi:10.1016/S1471-4906(01)02155-X. PMID 11929131.
- Monteiro RC, Van De Winkel JG (2003). "IgA Fc receptors". Annual Review of Immunology. 21: 177–204. doi:10.1146/annurev.immunol.21.120601.141011. PMID 12524384.
- Kremer EJ, Kalatzis V, Baker E, Callen DF, Sutherland GR, Maliszewski CR (April 1992). "The gene for the human IgA Fc receptor maps to 19q13.4". Human Genetics. 89 (1): 107–8. doi:10.1007/BF00207054. PMID 1577457. S2CID 22356303.
- Maliszewski CR, March CJ, Schoenborn MA, Gimpel S, Shen L (December 1990). "Expression cloning of a human Fc receptor for IgA". The Journal of Experimental Medicine. 172 (6): 1665–72. doi:10.1084/jem.172.6.1665. PMC 2188749. PMID 2258698.
- Pfefferkorn LC, Yeaman GR (October 1994). "Association of IgA-Fc receptors (Fc alpha R) with Fc epsilon RI gamma 2 subunits in U937 cells. Aggregation induces the tyrosine phosphorylation of gamma 2". Journal of Immunology. 153 (7): 3228–36. doi:10.4049/jimmunol.153.7.3228. PMID 7522255. S2CID 44693625.
- de Wit TP, Morton HC, Capel PJ, van de Winkel JG (August 1995). "Structure of the gene for the human myeloid IgA Fc receptor (CD89)". Journal of Immunology. 155 (3): 1203–9. doi:10.4049/jimmunol.155.3.1203. PMID 7636188. S2CID 12283570.
- Dürrbaum-Landmann I, Kaltenhäuser E, Flad HD, Ernst M (April 1994). "HIV-1 envelope protein gp120 affects phenotype and function of monocytes in vitro". Journal of Leukocyte Biology. 55 (4): 545–51. doi:10.1002/jlb.55.4.545. PMID 8145026. S2CID 44412688.
- Monteiro RC, Hostoffer RW, Cooper MD, Bonner JR, Gartland GL, Kubagawa H (October 1993). "Definition of immunoglobulin A receptors on eosinophils and their enhanced expression in allergic individuals". The Journal of Clinical Investigation. 92 (4): 1681–5. doi:10.1172/JCI116754. PMC 288327. PMID 8408621.
- Morton HC, Schiel AE, Janssen SW, van de Winkel JG (1996). "Alternatively spliced forms of the human myeloid Fc alpha receptor (CD89) in neutrophils". Immunogenetics. 43 (4): 246–7. doi:10.1007/s002510050057. PMID 8575829.
- Patry C, Sibille Y, Lehuen A, Monteiro RC (June 1996). "Identification of Fc alpha receptor (CD89) isoforms generated by alternative splicing that are differentially expressed between blood monocytes and alveolar macrophages". Journal of Immunology. 156 (11): 4442–8. doi:10.4049/jimmunol.156.11.4442. PMID 8666819. S2CID 45370016.
- Carayannopoulos L, Hexham JM, Capra JD (April 1996). "Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Calpha2 and Calpha3 in human IgA1". The Journal of Experimental Medicine. 183 (4): 1579–86. doi:10.1084/jem.183.4.1579. PMC 2192530. PMID 8666916.
- Pleass RJ, Andrews PD, Kerr MA, Woof JM (September 1996). "Alternative splicing of the human IgA Fc receptor CD89 in neutrophils and eosinophils". The Biochemical Journal. 318 ( Pt 3) (3): 771–7. doi:10.1042/bj3180771. PMC 1217685. PMID 8836118.
- Reterink TJ, Verweij CL, van Es LA, Daha MR (October 1996). "Alternative splicing of IgA Fc receptor (CD89) transcripts". Gene. 175 (1–2): 279–80. doi:10.1016/0378-1119(96)00152-7. PMID 8917112.
- van Dijk TB, Bracke M, Caldenhoven E, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP (December 1996). "Cloning and characterization of Fc alpha Rb, a novel Fc alpha receptor (CD89) isoform expressed in eosinophils and neutrophils". Blood. 88 (11): 4229–38. doi:10.1182/blood.V88.11.4229.bloodjournal88114229. PMID 8943858.
- Toyabe S, Kuwano Y, Takeda K, Uchiyama M, Abo T (November 1997). "IgA nephropathy-specific expression of the IgA Fc receptors (CD89) on blood phagocytic cells". Clinical and Experimental Immunology. 110 (2): 226–32. doi:10.1111/j.1365-2249.1997.tb08321.x. PMC 2265504. PMID 9367406.
- Gulle H, Samstag A, Eibl MM, Wolf HM (January 1998). "Physical and functional association of Fc alpha R with protein tyrosine kinase Lyn". Blood. 91 (2): 383–91. doi:10.1182/blood.V91.2.383. PMID 9427690.
External links
- CD89+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.