Bacterial DNA binding protein
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins.[1][2] Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature.[3] Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of bacteria (including cyanobacteria) and archaea, and are also encoded in the chloroplast genome of some algae.[4] The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control[5] is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R (or LMW5-AR).[6]
| Bac_DNA_binding | |||||||||
|---|---|---|---|---|---|---|---|---|---|
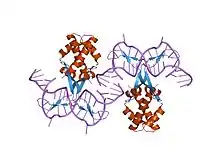 anabaena hu-dna cocrystal structure (ahu6) | |||||||||
| Identifiers | |||||||||
| Symbol | Bac_DNA_binding | ||||||||
| Pfam | PF00216 | ||||||||
| InterPro | IPR000119 | ||||||||
| PROSITE | PDOC00044 | ||||||||
| SCOP2 | 1hue / SCOPe / SUPFAM | ||||||||
| CDD | cd00591 | ||||||||
| |||||||||
This family is also found in a group of eukaryotes known as dinoflagellates. These dinoflagellate histone-like proteins replace histone in some dinoflagellates and package DNA into a liquid-crystalline state.[7]
History
Histone-like proteins are present in many Eubacteria, Cyanobacteria, and Archaebacteria. These proteins participate in all DNA-dependent functions; in these processes, bacterial DNA binding proteins have an architectural role, maintaining structural integrity as transcription, recombination, replication, or any other DNA-dependent process proceeds. Eukaryotic histones were first discovered through experiments in 0.4M NaCl. In these high salt concentrations, the eukaryotic histone protein is eluted from a DNA solution in which single stranded DNA is bound covalently to cellulose. Following elution, the protein readily binds DNA, indicating the protein's high affinity for DNA. Histone-like proteins were unknown to be present in bacteria until similarities between eukaryotic histones and the HU-protein were noted, particularly because of the abundancy, basicity, and small size of both of the proteins.[8] Upon further investigation, it was discovered that the amino acid composition of HU resembles that of eukaryotic histones, thus prompting further research into the exact function of bacterial DNA binding proteins and discoveries of other related proteins in bacteria.
Role in DNA replication
Research suggests that bacterial DNA binding protein has an important role during DNA replication; the protein is involved in stabilizing the lagging strand as well as interacting with DNA polymerase III. The role of single-stranded DNA binding (SSB) protein during DNA replication in Escherichia coli cells has been studied, specifically the interactions between SSB and the χ subunit of DNA polymerase III in environments of varying salt concentrations.[9]
In DNA replication at the lagging strand site, DNA polymerase III removes nucleotides individually from the DNA binding protein. An unstable SSB/DNA system would result in rapid disintegration of the SSB, which stalls DNA replication. Research has shown that the ssDNA is stabilized by the interaction of SSB and the χ subunit of DNA polymerase III in E. coli, thus preparing for replication by maintaining the correct conformation that increases the binding affinity of enzymes to ssDNA. Furthermore, binding of SSB to DNA polymerase III at the replication fork prevents dissociation of SSB, consequently increasing the efficiency of DNA polymerase III to synthesize a new DNA strand.
Examples
H-NS
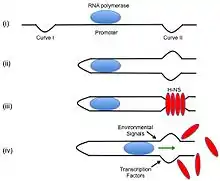
Initially, bacterial DNA binding proteins were thought to help stabilize bacterial DNA. Currently, many more functions of bacteria DNA binding proteins have been discovered, including the regulation of gene expression by histone-like nucleoid-structuring protein, H-NS.
H-NS is about 15.6 kDa and assists in the regulation of bacterial transcription in bacteria by repressing and activating certain genes. H-NS binds to DNA with an intrinsic curvature. In E. coli, H-NS binds to a P1 promoter decreasing rRNA production during stationary and slow growth periods. RNA polymerase and H-NS DNA binding protein have overlapping binding sites; it is thought that H-NS regulates rRNA production by acting on the transcription initiation site. It has been found that H-NS and RNA polymerase both bind to the P1 promoter and form a complex. When H-NS is bound with RNA Polymerase to the promoter region, there are structural differences in the DNA that are accessible.[11] It has also been found that H-NS can affect translation as well by binding to mRNA and causing its degradation.
HU
HU is a small (10 kDa[12]) bacterial DNA-binding protein, which structurally differs from a eukaryotic histone but functionally acts similarly to a histone by inducing negative supercoiling into circular DNA with the assistance of topoisomerase. The protein has been implicated in DNA replication, recombination, and repair. With an α-helical hydrophobic core and two positively charged β-ribbon arms, HU binds non-specifically to dsDNA with low affinity but binds to altered DNA—such as junctions, nicks, gaps, forks, and overhangs—with high affinity. The arms bind to the minor groove of DNA in low affinity states; in high affinity states, a component of the α-helical core interacts with the DNA as well. However, this protein's function is not solely confined to DNA; HU also binds to RNA and DNA-RNA hybrids with the same affinity as supercoiled DNA.[13]
Recent research has revealed that HU binds with high specificity to the mRNA of rpoS,[14] a transcript for the stress sigma factor of RNA polymerase, and stimulates translation of the protein. Additional to this RNA function, it was also demonstrated that HU binds DsrA, a small non-coding RNA that regulates transcription through repressing H-NS and stimulates translation through increasing expression of rpoS. These interactions suggest that HU has multiple influences on transcription and translation in bacterial cells.
IHF
Integration host factor, IHF, is not a nucleoid-associated protein only found in gram negative bacteria.[15] It is a 20 kDa heterodimer, composed of α and β subunits that bind to the sequence 5' - WATCAANNNNTTR - 3' and bends the DNA approximately 160 degrees.[16] The β arms of IHF have Proline residues that help stabilize the DNA kinks. These kinks can help compact DNA and allow for supercoiling. The mode of binding to DNA depends on environmental factors, such as the concentration of ions present. With a high concentration of KCl, there is weak DNA bending. It has been found that sharper DNA bending occurs when the concentration of KCl is less than 100 mM, and IHF is not concentrated.[17]
IHF was discovered as a necessary co-factor for recombination of λ phage into E.coli. In 2016 it was discovered that IHF also plays a key role in CRISPR type I and type II systems. It has a major role in allowing the Cas1-Cas2 complex to integrate new spacers into the CRISPR sequence. The bending of the DNA by IHF is thought to alter spacing in the DNA major and minor grooves, allowing the Cas1-Cas2 complex to make contact with the DNA bases.[18] This is a key function in the CRISPR system as it ensures that new spacers area always added at the beginning of the CRISPR sequence next to the leader sequence. This directing of integration by IHF ensures that spacers are added chronologically, allowing better protection against the most recent viral infection.[19]
Comparison
| DNA Binding Protein | Size | Structure | Binding Site | Effect |
|---|---|---|---|---|
| H-NS | 15.6 kDa | exists in dimers to physically prevent RNA polymerase from binding to promoter | binds to bent DNA, binds to P1 promoter in E. coli | regulation of gene expression |
| HU | 10 kDa | α-helical core and two positively charged β-ribbon arms | binds non-specifically to dsDNA, binds to DsrA, a small non-coding RNA that regulates transcription | induces negative supercoiling into circular DNA |
| IHF | 22 kDa | αβαβ hetrodimer | binds to specific sequences of DNA | creates kinks in DNA |
Implications and further research
The functions of bacterial DNA-binding proteins are not limited to DNA replication. Researchers have been investigating other pathways these proteins affect. The DNA-binding protein H-NS has been known to play roles in chromosome organization and gene regulation; however, recent studies have also confirmed their role in indirectly regulating flagella functions.[20] Some motility regulatory linkages that H-NS influences include the messenger molecule Cyclic di-GMP, the bio-film regulatory protein CsgD, and the sigma factors, σ(S) and σ(F). Further studies are aiming to characterize the ways this nucleoid-organizing protein affects the motility of the cell through other regulatory pathways.
Other researchers have used bacterial DNA-binding proteins to research Salmonella enterica serovar Typhimurium, in which the T6SS genes are activated from a macrophage infection. When S. Typhimurium infects, their efficiency can be improved through a sense-and-kill mechanism with T6SS H-NS silencing.[21] Assays are created that combine reporter fusions, electrophoretic mobility shift assays, DNase footprinting, and fluorescence microscopy to silence the T6SS gene cluster by the histone-like nucleoid structuring H-NS protein.
See also
References
- Drlica K, Rouviere-Yaniv J (September 1987). "Histonelike proteins of bacteria". Microbiological Reviews. 51 (3): 301–19. doi:10.1128/MMBR.51.3.301-319.1987. PMC 373113. PMID 3118156.
- Pettijohn DE (September 1988). "Histone-like proteins and bacterial chromosome structure". The Journal of Biological Chemistry. 263 (26): 12793–6. doi:10.1016/S0021-9258(18)37625-7. PMID 3047111.
- Griffiths, Anthony; Wessler, Susan; Carroll, Sean; Doebly, John. Introduction to Genetic Analysis (10 ed.). New York: W. H. Freeman and Company. pp. 428–429.
- Wang SL, Liu XQ (December 1991). "The plastid genome of Cryptomonas phi encodes an hsp70-like protein, a histone-like protein, and an acyl carrier protein". Proceedings of the National Academy of Sciences of the United States of America. 88 (23): 10783–7. Bibcode:1991PNAS...8810783W. doi:10.1073/pnas.88.23.10783. PMC 53015. PMID 1961745.
- Friedman DI (November 1988). "Integration host factor: a protein for all reasons" (PDF). Cell. 55 (4): 545–54. doi:10.1016/0092-8674(88)90213-9. hdl:2027.42/27063. PMID 2972385. S2CID 8548040.
- Neilan JG, Lu Z, Kutish GF, Sussman MD, Roberts PC, Yozawa T, Rock DL (March 1993). "An African swine fever virus gene with similarity to bacterial DNA binding proteins, bacterial integration host factors, and the Bacillus phage SPO1 transcription factor, TF1". Nucleic Acids Research. 21 (6): 1496. doi:10.1093/nar/21.6.1496. PMC 309344. PMID 8464748.
- Riaz, S; Sui, Z; Niaz, Z; Khan, S; Liu, Y; Liu, H (14 December 2018). "Distinctive Nuclear Features of Dinoflagellates with A Particular Focus on Histone and Histone-Replacement Proteins". Microorganisms. 6 (4): 128. doi:10.3390/microorganisms6040128. PMC 6313786. PMID 30558155.
- Drlica K, Rouviere-Yaniv J (September 1987). "Histonelike proteins of bacteria". Microbiological Reviews. 51 (3): 301–19. doi:10.1128/MMBR.51.3.301-319.1987. PMC 373113. PMID 3118156.
- Witte G, Urbanke C, Curth U (August 2003). "DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery". Nucleic Acids Research. 31 (15): 4434–40. doi:10.1093/nar/gkg498. PMC 169888. PMID 12888503.
- Dorman, Charles J; Deighan, Padraig (2003-04-01). "Regulation of gene expression by histone-like proteins in bacteria". Current Opinion in Genetics & Development. 13 (2): 179–184. doi:10.1016/S0959-437X(03)00025-X. PMID 12672495.
- Schröder O, Wagner R (May 2000). "The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex". Journal of Molecular Biology. 298 (5): 737–48. doi:10.1006/jmbi.2000.3708. PMID 10801345.
- Serban D, Arcineigas SF, Vorgias CE, Thomas GJ (April 2003). "Structure and dynamics of the DNA-binding protein HU of B. stearothermophilus investigated by Raman and ultraviolet-resonance Raman spectroscopy". Protein Science. 12 (4): 861–70. doi:10.1110/ps.0234103. PMC 2323852. PMID 12649443.
- Balandina A, Kamashev D, Rouviere-Yaniv J (August 2002). "The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids". The Journal of Biological Chemistry. 277 (31): 27622–8. doi:10.1074/jbc.M201978200. PMID 12006568.
- Balandina A, Claret L, Hengge-Aronis R, Rouviere-Yaniv J (February 2001). "The Escherichia coli histone-like protein HU regulates rpoS translation". Molecular Microbiology. 39 (4): 1069–79. doi:10.1046/j.1365-2958.2001.02305.x. PMID 11251825.
- Dillon SC, Dorman CJ (March 2010). "Bacterial nucleoid-associated proteins, nucleoid structure and gene expression". Nature Reviews. Microbiology. 8 (3): 185–95. doi:10.1038/nrmicro2261. PMID 20140026. S2CID 33103160.
- Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA (June 2016). "CRISPR Immunological Memory Requires a Host Factor for Specificity". Molecular Cell. 62 (6): 824–833. doi:10.1016/j.molcel.2016.04.027. PMID 27211867.
- Lin J, Chen H, Dröge P, Yan J (2012). "Physical organization of DNA by multiple non-specific DNA-binding modes of integration host factor (IHF)". PLOS ONE. 7 (11): e49885. Bibcode:2012PLoSO...749885L. doi:10.1371/journal.pone.0049885. PMC 3498176. PMID 23166787.
- Nuñez JK, Bai L, Harrington LB, Hinder TL, Doudna JA (June 2016). "CRISPR Immunological Memory Requires a Host Factor for Specificity". Molecular Cell. 62 (6): 824–833. doi:10.1016/j.molcel.2016.04.027. PMID 27211867.
- Sorek R, Lawrence CM, Wiedenheft B (2013). "CRISPR-mediated adaptive immune systems in bacteria and archaea". Annual Review of Biochemistry. 82 (1): 237–66. doi:10.1146/annurev-biochem-072911-172315. PMID 23495939.
- Kim EA, Blair DF (October 2015). "Function of the Histone-Like Protein H-NS in Motility of Escherichia coli: Multiple Regulatory Roles Rather than Direct Action at the Flagellar Motor". Journal of Bacteriology. 197 (19): 3110–20. doi:10.1128/JB.00309-15. PMC 4560294. PMID 26195595.
- Brunet YR, Khodr A, Logger L, Aussel L, Mignot T, Rimsky S, Cascales E (July 2015). "H-NS Silencing of the Salmonella Pathogenicity Island 6-Encoded Type VI Secretion System Limits Salmonella enterica Serovar Typhimurium Interbacterial Killing". Infection and Immunity. 83 (7): 2738–50. doi:10.1128/IAI.00198-15. PMC 4468533. PMID 25916986.