Ecdysteroid-phosphate phosphatase
Ecdysteroid-phosphate Phosphatase is the first structure of a steroid phosphate phosphotase containing alpha beta folds common to members of the two histidine (2H)-Phosphatase superfamily with strong homology to the Suppressor of T-cell receptor signaling-1 (Sts-1 PGM) protein. The putative EPPase PGM active site contains signature residues shared by 2H-phosphatase enzymes, including a conserved histidine (His80) that acts as a nucleophile during catalysis. The physiological substrate ecdysone 22-phosphate was modeled in a hydrophobic cavity close to the phosphate-binding site. EPPase PGM shows limited substrate specificity with an ability to hydrolyze steroid phosphates, the phospho-tyrosine (pTyr) substrate analogue para-nitrophenylphosphate ( pNPP) and pTyr-containing peptides and proteins. It has been shown that new protein tyrosine phosphatase (PTP) activity for EPPase. Also, EPPase and its closest homologues can be grouped into a distinct subfamily in the large 2H-Phosphatase superfamily of proteins.
Image
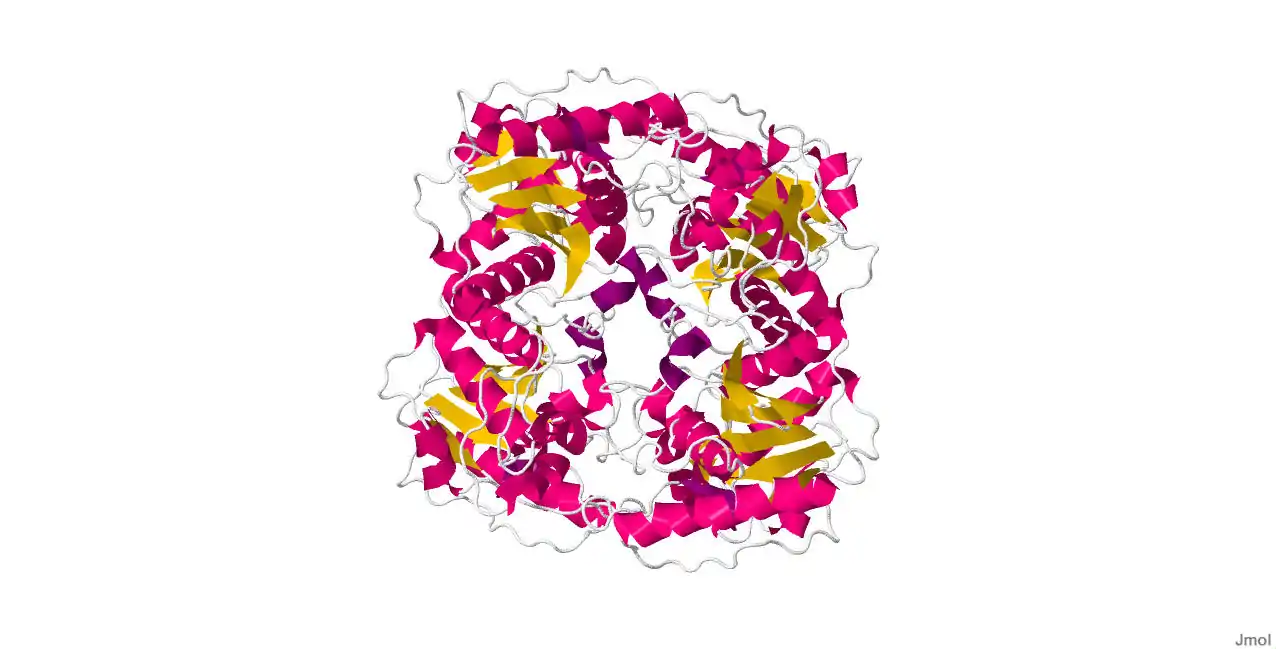
Concerning the sequence for EPPase, the following link will display where in the sequence alpha and beta strands can be found. http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=3C7T
Studies of EPPase
In some studies, the genes for EPPase were obtained from a silk moth while a strain of Escherichia coli served as the host for production of this receptor. A plasmid served as the vector for which this gene was implanted into the E.coli strain.
Structure
The structure of EPPase was obtained by x-ray diffraction and the following link provides another picture of the structure. https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=70358
References
- Chen, Y.; Carpino, N.; Nassar, N. "Protein Data Bank (PDB) "Crystal structure of the ecdysone phosphate phosphatase, EPPase, from Bombix mori in complex with tungstate"".