PETase
PETases are an esterase class of enzymes that catalyze the breakdown (via hydrolysis) of polyethylene terephthalate (PET) plastic to monomeric mono-2-hydroxyethyl terephthalate (MHET). The idealized chemical reaction is:
- (ethylene terephthalate)n + H2O → (ethylene terephthalate)n-1 + MHET,
| PETase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 3.1.1.101 | ||||||||
| Alt. names | PET hydrolase, poly(ethylene terephthalate) hydrolase | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
where n is the number of monomers in the polymer chain, though a trace amount of the PET breaks down instead to bis(2-hydroxyethyl) terephthalate (BHET).[1] PETases can also break down PEF-plastic (polyethylene-2,5-furandicarboxylate), which is a bioderived PET replacement, into the analogous MHEF. PETases can't catalyze the hydrolysis of aliphatic polyesters like polybutylene succinate or polylactic acid.[2]
Whereas the degradation of PET by natural (non-enzymatic) means will take hundreds of years, PETases can degrade it in a matter of days.[3]
History
The first PETase was discovered in 2016 from Ideonella sakaiensis strain 201-F6 bacteria found from sludge samples collected close to a Japanese PET bottle recycling site.[1][4] There were other types of hydrolases previously known to degrade PET,[2] including lipases, esterases, and cutinases.[5] For comparison, enzymes that degrade polyester have been known to exist at least as far back as 1975 (in the case of α-chymotrypsin)[6] and 1977 (lipase).[7]
PET plastic came into widespread use in the 1970s and it has been suggested that PETases in bacteria evolved only recently.[2] PETase may have had past enzymatic activity associated with degradation of a waxy coating on plants.[8]
Structure
As of April 2019, there were 17 known three-dimensional crystal structures of PETases: 6QGC, 6ILX, 6ILW, 5YFE, 6EQD, 6EQE, 6EQF, 6EQG, 6EQH, 6ANE, 5XJH, 5YNS, 5XFY, 5XFZ, 5XG0, 5XH2 and 5XH3.
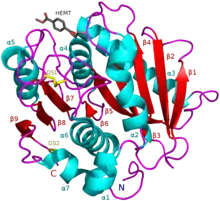
PETase exhibits shared qualities with both lipases and cutinases in that it possesses an α/β-hydrolase fold; although, the active-site cleft observed in PETase is more open than in cutinases.[2] The Ideonella sakaiensis PETase is similar to dienelactone hydrolase, according to Pfam. According to ESTHER, it falls into the Polyesterase-lipase-cutinase family.
There are approximately 69 PETase-like enzymes comprising a variety of diverse organisms, and there are two classifications of these enzymes including type I and type II. It is suggested that 57 enzymes fall into the type I category whereas the rest fall into the type II group, including the PETase enzyme found in the Ideonella sakaiensis. Within all 69 PETase-like enzymes, there exists the same three residues within the active site, suggesting that the catalytic mechanism is the same in all forms of PETase-like enzymes.[9]
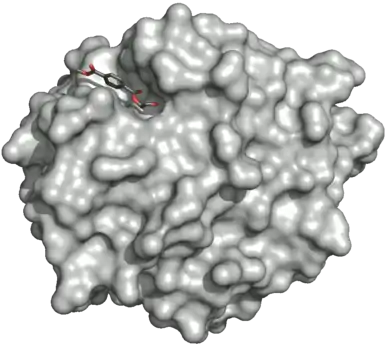 Surface of the PETase double mutant (R103G and S131A) with HEMT (1-(2-hydroxyethyl) 4-methyl terephthalate) bound to its active site. HEMT is an analogue of MHET, and has an additional methanol esterified to it. PDBID: 5XH3.
Surface of the PETase double mutant (R103G and S131A) with HEMT (1-(2-hydroxyethyl) 4-methyl terephthalate) bound to its active site. HEMT is an analogue of MHET, and has an additional methanol esterified to it. PDBID: 5XH3.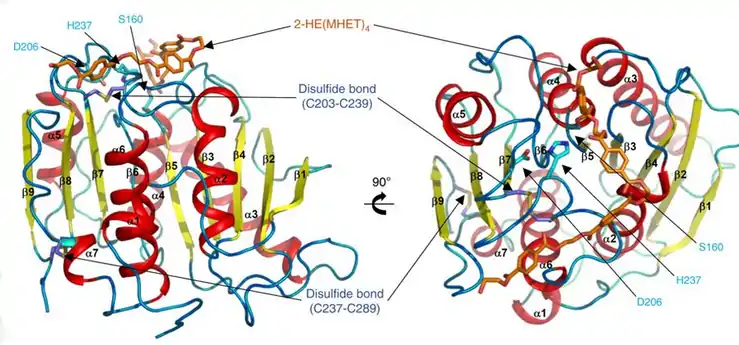 Ribbon diagram of PETase with three residues Ser160, Asp206, and His237. The catalytic triad is represented by cyan-colored sticks. The active site is shown in orange to represent stimulation by a 2-HE(MHET)4 molecule.[9]
Ribbon diagram of PETase with three residues Ser160, Asp206, and His237. The catalytic triad is represented by cyan-colored sticks. The active site is shown in orange to represent stimulation by a 2-HE(MHET)4 molecule.[9]
Mutations
In 2018 scientists from the University of Portsmouth with the collaboration of the National Renewable Energy Laboratory of the United States Department of Energy developed a mutant of this PETase that degrades PET faster than the one in its natural state. In this study it was also shown that PETases can degrade polyethylene 2,5-furandicarboxylate (PEF).[2][10]
Studies have demonstrated that the β1-β2 connecting loop located far from the active site of PETase of I. sakaiensis (IsPETase) structure is very flexible it is suggested that it can be considered for mutagenesis to increase the thermal stability of IsPETase.[10]
Biological pathway

In I. sakaiensis, the resultant MHET is further broken down by the action of MHETase enzyme to terephthalic acid and ethylene glycol.[1] Laboratory experiments showed that chimeric proteins that artificially link a MHETase and a PETase outperform similar mixtures of free enzymes.[12]
See also
- Organisms breaking down plastic
- Galleria mellonella, a caterpillar that can digest polyethylene.
- Aspergillus tubingensis, a fungus that can digest polyurethane.
- Pestalotiopsis microspora, an endophytic fungus species able to break down polyurethane.
- cutinase, an esterase enzyme of similar geometric shape
References
- Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, et al. (March 2016). "A bacterium that degrades and assimilates poly(ethylene terephthalate)". Science. 351 (6278): 1196–9. doi:10.1126/science.aad6359. PMID 26965627. S2CID 31146235.
- "Discovery of a Bacterium that Degrades and Assimilates Poly(ethylene terephthalate) could Serve as a Degradation and/or Fermentation Platform for Biological Recycling of PET Waste Products" (PDF). Kyoto Institute of Technology (Press release). 2016-03-30.
- Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, et al. (May 2018). "Characterization and engineering of a plastic-degrading aromatic polyesterase". Proceedings of the National Academy of Sciences of the United States of America. 115 (19): E4350–E4357. doi:10.1073/pnas.1718804115. PMC 5948967. PMID 29666242.
- Dockrill, Peter. "Scientists Have Accidentally Created a Mutant Enzyme That Eats Plastic Waste". ScienceAlert. Retrieved 2018-11-27.
- Tanasupawat S, Takehana T, Yoshida S, Hiraga K, Oda K (August 2016). "Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate)". International Journal of Systematic and Evolutionary Microbiology. 66 (8): 2813–8. doi:10.1099/ijsem.0.001058. PMID 27045688.
- Han X, Liu W, Huang JW, Ma J, Zheng Y, Ko TP, et al. (December 2017). "Structural insight into catalytic mechanism of PET hydrolase". Nature Communications. 8 (1): 2106. doi:10.1038/s41467-017-02255-z. PMC 5727383. PMID 29235460.
- Tabushi I, Yamada H, Matsuzaki H, Furukawa J (August 1975). "Polyester readily hydrolyzable by chymotrypsin". Journal of Polymer Science: Polymer Letters Edition. 13 (8): 447–450. doi:10.1002/pol.1975.130130801.
- Tokiwa Y, Suzuki T (November 1977). "Hydrolysis of polyesters by lipases". Nature. 270 (5632): 76–8. doi:10.1038/270076a0. PMID 927523. S2CID 4145159.
- "Lab 'Accident' Becomes Mutant Enzyme That Devours Plastic". Live Science. Retrieved 2018-11-27.
- Joo S, Cho IJ, Seo H, Son HF, Sagong HY, Shin TJ, et al. (January 2018). "Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation". Nature Communications. 9 (1): 382. doi:10.1038/s41467-018-02881-1. PMC 5785972. PMID 29374183.
- da Costa, Clauber. "Assessment of the PETase conformational changes induced by poly(ethylene terephthalate) binding". Proteins: Struture, Functions, and Bioinformatics.
- Allison Chan (2016). "The Future of Bacteria Cleaning Our Plastic Waste" (PDF).
- Knott BC, Erickson E, Allen MD, Gado JE, Graham R, Kearns FL, et al. (Oct 2020). "Characterization and engineering of a two-enzyme system for plastics depolymerization". Proc Natl Acad Sci U S A. 117 (41): 25476–25485. doi:10.1073/pnas.2006753117. PMC 7568301. PMID 32989159.