Genetic history of the Indigenous peoples of the Americas
The genetic history of the Indigenous peoples of the Americas is divided into two distinct periods: the initial peopling of the Americas during about 20,000 to 14,000 years ago (20–14 kya), and European contact, after about 500 years ago.[1][2] The first period of Indigenous American genetic history is the determinant factor for the number of genetic lineages, zygosity mutations and founding haplotypes present in today's Indigenous American populations.[3]
Indigenous American populations descend from an Ancient Paleo-Siberian population, itself a combination of an Ancient East Asian lineage which diverged from other East Asian peoples prior to the Last Glacial Maximum, between 36,000 and 25,000 years ago, and subsequently migrated into Siberia, where they merged with a primarily West Eurasian (Ancient North Eurasians), deeply related to European hunter-gatherers. They later dispersed throughout the Americas after about 16,000 years ago (exceptions being the Na-Dene and Eskimo–Aleut speaking groups, which are derived partially from Siberian populations which entered the Americas at a later time).[4][5]
Analyses of genetics among Indigenous American and Siberian populations have been used to argue for early isolation of founding populations on Beringia[6] and for later, more rapid migration from Siberia through Beringia into the New World.[7] The microsatellite diversity and distributions of the Y lineage specific to South America indicates that certain Indigenous American populations have been isolated since the initial peopling of the region.[8] The Na-Dene, Inuit and Native Alaskan populations exhibit Haplogroup Q-M242; however, they are distinct from other Indigenous Americans with various mtDNA and atDNA mutations.[9][10][11] This suggests that the peoples who first settled in the northern extremes of North America and Greenland derived from later migrant populations than those who penetrated farther south in the Americas.[12][13] Linguists and biologists have reached a similar conclusion based on analysis of Indigenous American language groups and ABO blood group system distributions.[14][15][16][17]
Autosomal DNA

Genetic diversity and population structure in the American landmass is also measured using autosomal (atDNA) micro-satellite markers genotyped; sampled from North, Central, and South America and analyzed against similar data available from other Indigenous populations worldwide.[18][19] The Indigenous American populations show a lesser genetic diversity than populations from other continental regions.[19] Observed is a decreasing genetic diversity as geographic distance from the Bering Strait occurs, as well as a decreasing genetic similarity to Siberian populations from Alaska (the genetic entry point).[18][19] Also observed is evidence of a greater level of diversity and lesser level of population structure in western South America compared to eastern South America.[18][19] There is a relative lack of differentiation between Mesoamerican and Andean populations, a scenario that implies that coastal routes (in this case along the coast of the Pacific Ocean) were easier for migrating peoples (more genetic contributors) to traverse in comparison with inland routes.[18]
The over-all pattern that is emerging suggests that the Americas were colonized by a small number of individuals (effective size of about 70), which grew by many orders of magnitude over 800 – 1000 years.[20][21] The data also shows that there have been genetic exchanges between Asia, the Arctic, and Greenland since the initial peopling of the Americas.[21][22]
According to an autosomal genetic study from 2012,[23] Indigenous Americans descend from at least three main migrant waves from East Asia. Most of it is traced back to a single ancestral population, called 'First Americans'. However, those who speak Inuit languages from the Arctic inherited almost half of their ancestry from a second East Asian migrant wave. And those who speak Na-Dene, on the other hand, inherited a tenth of their ancestry from a third migrant wave. The initial settling of the Americas was followed by a rapid expansion southwards along the west coast, with little gene flow later, especially in South America. One exception to this are the Chibcha speakers of Colombia, whose ancestry comes from both North and South America.[23]
In 2014, the autosomal DNA of a 12,500+ year old infant from Montana was sequenced.[24] The DNA was taken from a skeleton referred to as Anzick-1, found in close association with several Clovis artifacts. Comparisons showed strong affinities with DNA from Siberian sites, and virtually ruled out that particular individual had any close affinity with European sources (the "Solutrean hypothesis"). The DNA also showed strong affinities with all existing Indigenous American populations, which indicated that all of them derive from an ancient population that lived in or near Siberia.[25]
Linguistic studies have reinforced genetic studies, with relationships between languages found among those spoken in Siberia and those spoken in the Americas.[26]
Two 2015 autosomal DNA genetic studies confirmed the Siberian origins of the Indigenous peoples of the Americas. However an ancient signal of shared ancestry with Australasians (Indigenous peoples of Australia, Melanesia and the Andaman Islands) was detected among the Indigenous peoples of the Amazon region. The migration coming out of Siberia would have happened 23,000 years ago.[27][28][29]
A 2018 study analysed 11,500 BCE old Indigenous samples. The genetic evidence suggests that all Indigenous Americans ultimately descended from a single founding population that initially split from a Basal-East Asian source population in Mainland Southeast Asia around 36,000 years ago, the same time at which the proper Jōmon people divided from Basal-East Asians, either together with Ancestral Indigenous Americans or during a separate expansion wave. The authors also provided evidence that the basal northern and southern Indigenous American branches, to which all other Indigenous peoples belong, diverged around 16,000 years ago.[4][30] An Indigenous American sample from 16,000 BCE in Idaho, which is craniometrically similar to modern Indigenous Americans as well as Paleosiberians, was found to have been largely East-Eurasian genetically, and showed high affinity with contemporary East Asians, as well as Jōmon period samples of Japan, confirming that Ancestral Indigenous Americans split from an East-Eurasian source population somewhere in eastern Siberia.[31]
A study published in the Nature journal in 2018 concluded that Indigenous Americans descended from a single founding population which initially divided from East Asians about ~36,000 BCE, with gene flow between Ancestral Indigenous Americans and Siberians persisting until ~25,000 BCE, before becoming isolated in the Americas at ~22,000 BCE. Northern and Southern American Indigenous sub-populations split from each other at ~17,500 BCE. There is also some evidence for a back-migration from the Americas into Siberia after ~11,500 BCE.[4]
A study published in the Cell journal in 2019, analysed 49 ancient Indigenous American samples from all over North and South America, and concluded that all Indigenous American populations descended from a single ancestral source population which divided from Siberians and East Asians, and gave rise to the Ancestral Indigenous Americans, which later diverged into the various Indigenous groups. The authors further dismissed previous claims for the possibility of two distinct population groups among the peopling of the Americas. Both, Northern and Southern Indigenous Americans are closest to each other, and do not show evidence of admixture with hypothetical previous populations.[32]
A review article published in the Nature journal in 2021, which summarized the results of previous genomic studies, similarly concluded that all Indigenous Americans descended from the movement of people from Northeast Asia into the Americas. These Ancestral Americans, once south of the continental ice sheets, spread and expanded rapidly, and branched into multiple groups, which later gave rise to the major subgroups of Indigenous American populations. The study also dismissed the existence, inferred from craniometric data, of a hypothetical distinct non-Indigenous American population (suggested to have been related to Indigenous Australians and Papuans), sometimes called "Paleoamerican".[33][34]
Overall, the 'Ancestral Native Americans' formed from an 'Ancient Paleo-Siberian' lineage which formed from the admixture between East Asian people and a distinct Paleolithic Siberian population known as Ancient North Eurasians, closer related to modern Europeans, giving rise to both Indigenous peoples of Siberia and Native Americans.[35]
Y-chromosome DNA
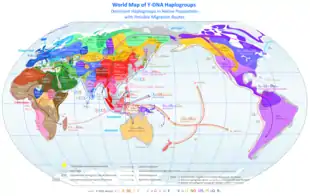
A "Central Siberian" origin has been postulated for the paternal lineage of the source populations of the original migration into the Americas.[36]
Membership in haplogroups Q and C3b implies Indigenous American patrilineal descent.[37]
The micro-satellite diversity and distribution of a Y lineage specific to South America suggest that certain Indigenous American populations became isolated after the initial colonization of their regions.[8] The Na-Dene, Inuit, and Native Alaskan populations exhibit haplogroup Q (Y-DNA) mutations, but are distinct from other Indigenous Americans with various mtDNA and autosomal DNA (atDNA) mutations.[9][38][39] This suggests that the earliest migrants into the northern extremes of North America and Greenland derived from later migrant populations.[40][41]
Haplogroup Q
.png.webp)
Q-M242 (mutational name) is the defining (SNP) of Haplogroup Q (Y-DNA) (phylogenetic name).[43][44] In Eurasia, haplogroup Q is found among Indigenous Siberian populations, such as the modern Chukchi and Koryak peoples, as well as some Southeast Asians, such as the Dayak people. In particular, two groups exhibit large concentrations of the Q-M242 mutation, the Ket (93.8%) and the Selkup (66.4%) peoples.[45] The Ket are thought to be the only survivors of ancient wanderers living in Siberia.[46] Their population size is very small; there are fewer than 1,500 Ket in Russia.2002[20] The Selkup have a slightly larger population size than the Ket, with approximately 4,250 individuals.[47]
Starting the Paleo-Indigenous American period, a migration to the Americas across the Bering Strait (Beringia) by a small population carrying the Q-M242 mutation occurred.[10] A member of this initial population underwent a mutation, which defines its descendant population, known by the Q-M3 (SNP) mutation.[48] These descendants migrated all over the Americas.[43]
Haplogroup Q-M3 is defined by the presence of the rs3894 (M3) (SNP).[1][20][49] The Q-M3 mutation is roughly 15,000 years old as that is when the initial migration of Paleo-Indigenous Americans into the Americas occurred.[50][51] Q-M3 is the predominant haplotype in the Americas, at a rate of 83% in South American populations,[8] 50% in the Na-Dene populations, and in North American Eskimo-Aleut populations at about 46%.[45] With minimal back-migration of Q-M3 in Eurasia, the mutation likely evolved in east-Beringia, or more specifically the Seward Peninsula or western Alaskan interior. The Beringia land mass began submerging, cutting off land routes.[45][52][18]
Since the discovery of Q-M3, several subclades of M3-bearing populations have been discovered. An example is in South America, where some populations have a high prevalence of (SNP) M19, which defines subclade Q-M19.[8] M19 has been detected in (59%) of Amazonian Ticuna men and in (10%) of Wayuu men.[8] Subclade M19 appears to be unique to South American Indigenous peoples, arising 5,000 to 10,000 years ago.[8] This suggests that population isolation, and perhaps even the establishment of tribal groups, began soon after migration into the South American areas.[20][53] Other American subclades include Q-L54, Q-Z780, Q-MEH2, Q-SA01, and Q-M346 lineages. In Canada, two other lineages have been found. These are Q-P89.1 and Q-NWT01.
Haplogroup R1
Haplogroup R1 is the second most common Y-DNA haplogroup found among Indigenous Americans after Y-DNA haplogroup Q.[54]
Initially, there was debate about the origin of haplogroup R1b in Native Americans. Two early studies suggested that this haplogroup could have been one of the founding Siberian lineages of Native Americans, however this is now considered unlikely, because the R1b lineages commonly found in Native Americans are in most cases identical to those in western Europeans, and its highest concentration is found among a variety of culturally unaffiliated tribes, in eastern North America.[55]
Thus, according to several authors, R1b was most likely introduced through admixture during the post-1492 European settlement of North America.[56][57][58]
R1 (M173) is found predominantly in North American groups like the Ojibwe (50-79%), Seminole (50%), Sioux (50%), Cherokee (47%), Dogrib (40%) and Tohono O'odham (Papago) (38%). Its highest frequency is found in northeastern North America, and declines in frequency from east to west. In southwestern Native American tribes the frequency of this haplogroup is as low as 4%.[54][59]
Haplogroup C-P39
.PNG.webp)
Haplogroup C-M217 is found mainly in Indigenous Siberians, Mongolians, and Kazakhs. Haplogroup C-M217 is the most widespread and frequently occurring branch of the greater (Y-DNA) haplogroup C-M130. Haplogroup C-M217 descendant C-P39 is most commonly found in today's Na-Dene speakers, with the greatest frequency found among the Athabaskans at 42%, and at lesser frequencies in some other Indigenous American groups.[10] This distinct and isolated branch C-P39 includes almost all the Haplogroup C-M217 Y-chromosomes found among all Indigenous peoples of the Americas.[61]
Some researchers feel that this may indicate that the Na-Dene migration occurred from the Russian Far East after the initial Paleo-Indigenous American colonization, but prior to modern Inuit, Inupiat and Yupik expansions.[10][9][62]
In addition to in Na-Dene peoples, haplogroup C-P39 (C2b1a1a) is also found among other Indigenous Americans such as Algonquian- and Siouan-speaking populations.[63][64] C-M217 is found among the Wayuu people of Colombia and Venezuela.[63][64]
Data
Listed here are notable Indigenous peoples of the Americas by human Y-chromosome DNA haplogroups based on relevant studies. The samples are taken from individuals identified with the ethnic and linguistic designations in the first two columns, the fourth column (n) is the sample size studied, and the other columns give the percentage of the particular haplogroup.
Mitochondrial DNA
The common occurrence of the mtDNA Haplogroups A, B, C, and D among eastern Asian and Indigenous American populations has long been recognized, along with the presence of Haplogroup X.[68] As a whole, the greatest frequency of the four Indigenous American associated haplogroups occurs in the Altai-Baikal region of southern Siberia.[69] Some subclades of C and D closer to the Indigenous American subclades occur among Mongolian, Amur, Japanese, Korean, and Ainu populations.[68][70] A 2023 DNA study found that "[i]n addition to previously described ancestral sources in Siberia, Australo-Melanesia, and Southeast Asia, ... northern coastal China also contributed to the gene pool of Native Americans" as well as that of Japanese people.[71]
.PNG.webp)
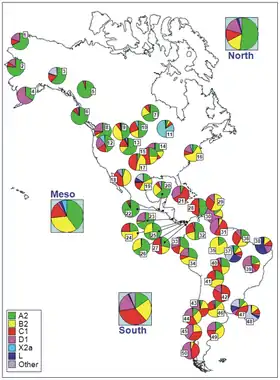
When studying human mitochondrial DNA haplogroup, the results indicated that Indigenous American haplogroups, including haplogroup X, are part of a single founding East Asian population. It also indicates that the distribution of mtDNA haplogroups and the levels of sequence divergence among linguistically similar groups were the result of multiple preceding migrations from Bering Straits populations.[72] All Indigenous American mtDNA can be traced back to five haplogroups, A, B, C, D and X.[73][74] More specifically, Indigenous American mtDNA belongs to sub-haplogroups A2, B2, C1b, C1c, C1d, D1, and X2a (with minor groups C4c, D2a, and D4h3a).[6][72] This suggests that 95% of Indigenous American mtDNA is descended from a minimal genetic founding female population, comprising sub-haplogroups A2, B2, C1b, C1c, C1d, and D1.[73] The remaining 5% is composed of the X2a, D2a, C4c, and D4h3a sub-haplogroups.[72][73]
X is one of the five mtDNA haplogroups found in Indigenous Americans. Native Americans mostly belong to the X2a clade, which has never been found in the Old World.[75] According to Jennifer Raff, X2a probably originated in the same Siberian population as the other four founding maternal lineages.[76]
Haplogroup X genetic sequences diverged about 20,000 to 30,000 years ago to give two sub-groups, X1 and X2. X2's subclade X2a occurs only at a frequency of about 3% for the total current Indigenous population of the Americas.[20] However, X2a is a major mtDNA subclade in North America; among the Algonquian peoples, it comprises up to 25% of mtDNA types.[1][77] It is also present in lower percentages to the west and south of this area — among the Sioux (15%), the Nuu-chah-nulth (11%–13%), the Navajo (7%), and the Yakama (5%).[78] The predominant theory for sub-haplogroup X2a's appearance in North America is migration along with A, B, C, and D mtDNA groups, from a source in the Altai Mountains of central Asia.[79][80][81][82] Haplotype X6 was present in the Tarahumara 1.8% (1/53) and Huichol 20% (3/15)[83]
Sequencing of the mitochondrial genome from Paleo-Eskimo remains (3,500 years old) are distinct from modern Indigenous Americans, falling within sub-haplogroup D2a1, a group observed among today's Aleutian Islanders, the Aleut and Siberian Yupik populations.[84] This suggests that the colonizers of the far north, and subsequently Greenland, originated from later coastal populations.[84] Then a genetic exchange in the northern extremes introduced by the Thule people (proto-Inuit) approximately 800–1,000 years ago began.[39][85] These final Pre-Columbian migrants introduced haplogroups A2a and A2b to the existing Paleo-Eskimo populations of Canada and Greenland, culminating in the modern Inuit.[39][85]
A route through Beringia is seen as more likely than the Solutrean hypothesis.[86] An abstract in a 2012 issue of the "American Journal of Physical Anthropology" states that "The similarities in ages and geographical distributions for C4c and the previously analyzed X2a lineage provide support to the scenario of a dual origin for Paleo-Indigenous Americans. Taking into account that C4c is deeply rooted in the Asian portion of the mtDNA phylogeny and is indubitably of Asian origin, the finding that C4c and X2a are characterized by parallel genetic histories definitively dismisses the controversial hypothesis of an Atlantic glacial entry route into North America."[87]
Another study, also focused on the mtDNA (that which is inherited through only the maternal line),[6] revealed that the Indigenous people of the Americas have their maternal ancestry traced back to a few founding lineages from East Asia, which would have arrived via the Bering strait. According to this study, it is probable that the ancestors of the Indigenous Americans would have remained for a time in the region of the Bering Strait, after which there would have been a rapid movement of settling of the Americas, taking the founding lineages to South America.
According to a 2016 study, focused on mtDNA lineages, "a small population entered the Americas via a coastal route around 16.0 ka, following previous isolation in eastern Beringia for ~2.4 to 9 thousand years after separation from eastern Siberian populations. Following a rapid movement throughout the Americas, limited gene flow in South America resulted in a marked phylogeographic structure of populations, which persisted through time. All of the ancient mitochondrial lineages detected in this study were absent from modern data sets, suggesting a high extinction rate. To investigate this further, we applied a novel principal components multiple logistic regression test to Bayesian serial coalescent simulations. The analysis supported a scenario in which European colonization caused a substantial loss of pre-Columbian lineages".[88]
Genetic admixture
Ancient Beringians

Recent archaeological findings in Alaska have shed light on the existence of a previously unknown Indigenous American population that has been academically named "Ancient Beringians."[89] Although it is popularly agreed among archeologists that early settlers had crossed into Alaska from Russia through the Bering Strait land bridge, the issue of whether or not there was one founding group or several waves of migration is a controversial and prevalent debate among academics in the field today. In 2018, the sequenced DNA of a Indigenous girl, whose remains were found at the Sun River archaeological site in Alaska in 2013, proved not to match the two recognized branches of Indigenous Americans and instead belonged to the early population of Ancient Beringians.[90] This breakthrough is said to be the first direct genomic evidence that there was potentially only one wave of migration in the Americas that occurred, with genetic branching and division transpiring after the fact. The migration wave is estimated to have emerged about 20,000 years ago.[89] The Ancient Beringians are said to be a common ancestral group among contemporary Indigenous American populations today, which differs in results collected from previous research that suggests that modern populations are descendants of either Northern and Southern branches.[89] Experts were also able to use wider genetic evidence to establish that the split between the Northern and Southern American branches of civilization from the Ancient Beringians in Alaska only occurred about 17,000 and 14,000 years,[23] further challenging the concept of multiple migration waves occurring during the very first stages of settlement.
Genetic evidence for Paleo-Indigenous Americans consists of the presence of apparent admixture of archaic Sundadont lineages to the remote populations in the South American rain forest, and in the genetics of Patagonians-Fuegians.[91][92]
Nomatto et al. (2009) proposed migration into Beringia occurred between 40k and 30k cal years BP, with a pre-LGM migration into the Americas followed by isolation of the northern population following closure of the ice-free corridor.[93]
A 2016 genetic study of Indigenous peoples of the Amazonian region of Brazil (by Skoglund and Reich) showed evidence of admixture from a separate lineage of an otherwise unknown ancient people. This ancient group appears to be related to modern day "Australasian" peoples (i.e. Aboriginal Australians and Melanesians). This "Ghost population" was found in speakers of Tupian languages. They provisionally named this ancient group; "Population Y", after Ypykuéra, "which means 'ancestor' in the Tupi language family".[29] A 2021 genetic study dismissed the existence of an hypothetical Australasian component among Indigenous Americans. The signal of the hypothetical Australasian component, can also be reproduced using the Basal-East Asian Tianyuan man sample, and thus does not represent "real Australasian affinity". The authors explained that the previous claims of possibly Australasian ancestry were based on a misinterpreted genetic echo, which was revealed to represent early East-Eurasian gene flow (represented by the 40,000 BCE old Tianyuan sample) into Aboriginal Australians and Papuans, which was lost in modern East Asians.[33][34]
Archaeological evidence for pre-LGM human presence in the Americas was first presented in the 1970s.[94][95] notably the "Luzia Woman" skull found in Brazil.[96][97][98]
Old world
Substantial racial admixture has taken place during and since the European colonization of the Americas.[99][100]
South and Central America
In Latin America in particular, significant racial admixture took place between the Indigenous American population, the European-descended colonial population, and the Sub-Saharan African populations imported as slaves. From about 1700, a Latin American terminology developed to refer to the various combinations of mixed racial descent produced by this.[101]
Many individuals who self-identify as one race exhibit genetic evidence of a multiracial ancestry.[102] The European conquest of South and Central America, beginning in the late 15th century, was initially executed by male soldiers and sailors from the Iberian Peninsula (Spain and Portugal).[103] The new soldier-settlers fathered children with Indigenous American women and later with African slaves.[104] These mixed-race children were generally identified by the Spanish colonist and Portuguese colonist as "Castas".[105]
North America
The North American fur trade during the 16th century brought many European men, from France, Ireland, and Great Britain, who married Indigenous North American women.[106] In the areas where these peoples formed communities, and developed a unique, syncretic culture, their children became known as "Métis" or "Bois-Brûlés" by the French colonists. In some contexts these peoples have also been referred to as "mixed-bloods", or "country-born" by the English and Scottish colonists.[107]
Native Americans in the United States are defined by citizenship, culture, and familial relationships, not race.[108][109] Having never defined Native American identity as racial,[108] historically, Native Americans have commonly practiced what mainstream society defines as interracial marriage, which has affected racial ideas of blood quantum.[110]
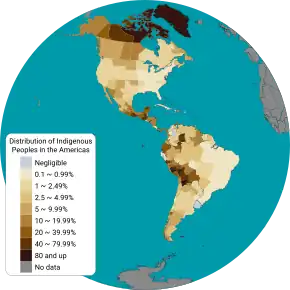
In the United States 2010 census, nearly 3 million people indicated that their race was Indigenous American (including Alaskan Native).[111] This is based on self-identification, as the census does not require documentation of this belief. Especially numerous was the self-identification of Cherokee ethnic origin,[112] a phenomenon dubbed the "Cherokee Syndrome", where some Americans believe they have a "long-lost Cherokee ancestor" without being able to identify any Cherokee or Native American people in their family tree or among their living relatives.[113][114] This cultivation of an opportunistic ethnic identity is related to the "prestige" non-Natives may associate with Indigenous American ancestry, having never experienced any of the attendant hardships or oppression.[115] In the Eastern United States, in particular, pretendians are common.[115][116]
Some tribes have adopted blood quantum requirements, or Certificates of Degree of Indian Blood, and practice disenrollment of tribal members unable to prove they are the child of an enrolled tribal member. In these cases, the tribe may demand a paternity test. For some, this has become a contentious issue in Native American reservation politics.[117]
European diseases and genetic modification
A team led by Ripan Malhi, an anthropologist at the University of Illinois in Urbana, conducted a study where they used a scientific technique known as whole exome sequencing to test immune-related gene variants within Indigenous Americans.[118] Through analyzing ancient and modern Indigenous DNA, it was found that HLA-DQA1, a variant gene that codes for protein in charge of differentiating between healthy cells from invading viruses and bacteria were present in nearly 100% of ancient remains but only 36% in modern Indigenous Americans.[118] These finding suggest that European-borne epidemics such as smallpox altered the disease landscape of the Americas, leaving survivors of these outbreaks less likely to carry variants like HLA-DQA1. This made them less able to cope with new diseases. The change in genetic makeup is measured by scientists to have occurred around 175 years ago, during a time when the smallpox epidemic was ranging through the Americas.
Blood groups
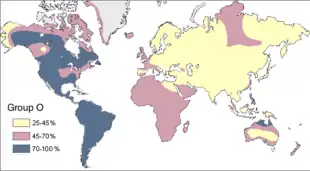
Prior to the 1952 confirmation of DNA as the hereditary material by Alfred Hershey and Martha Chase, scientists used blood proteins to study human genetic variation.[119][120] The ABO blood group system is widely credited to have been discovered by the Austrian Karl Landsteiner, who found three different blood types in 1900.[121] Blood groups are inherited from both parents. The ABO blood type is controlled by a single gene (the ABO gene) with three alleles: i, IA, and IB.[122]
Research by Ludwik and Hanka Herschfeld during World War I found that the frequencies of blood groups A, B and O differed greatly from region to region.[120] The "O" blood type (usually resulting from the absence of both A and B alleles) is very common around the world, with a rate of 63% in all human populations.[123] Type "O" is the primary blood type among the Indigenous populations of the Americas, particularly within Central and South American populations, with a frequency of nearly 100%.[123] In Indigenous North American populations the frequency of type "A" ranges from 16% to 82%.[123] This suggests again that the initial Indigenous Americans evolved from an isolated population with a minimal number of individuals.[124][125]
The standard explanation for such a high population of Indigenous Americans with blood type O is genetic drift. Because the ancestral population of Indigenous Americans was numerically small, blood type diversity could have been reduced from generation to generation by the founder effect.[126] Other related explanations include the Bottleneck explanation which states that there were high frequencies of blood type A and B among Indigenous Americans but severe population decline during the 1500s and 1600s caused by the introduction of disease from Europe resulted in the massive death toll of those with blood types A and B. Coincidentally, a large amount of the survivors were type O.[126]
| PEOPLE GROUP | O (%) | A (%) | B (%) | AB (%) |
|---|---|---|---|---|
| Blackfoot Confederacy (Indigenous North American) | 17 | 82 | 0 | 1 |
| Bororo (Brazil) | 100 | 0 | 0 | 0 |
| Eskimos (Alaska) | 38 | 44 | 13 | 5 |
| Inuit (Eastern Canada & Greenland) | 54 | 36 | 23 | 8 |
| Hawaiians (Polynesians, non-Indigenous American) | 37 | 61 | 2 | 1 |
| Indigenous North Americans (as a whole Native Nations/First Nations) | 79 | 16 | 4 | 1 |
| Maya (modern) | 98 | 1 | 1 | 1 |
| Navajo | 73 | 27 | 0 | 0 |
| Peru | 100 | 0 | 0 | 0 |
The Dia antigen of the Diego antigen system has been found only in Indigenous peoples of the Americas and East Asians, and in people with some ancestry from those groups. The frequency of the Dia antigen in various groups of Indigenous peoples of the Americas ranges from almost 50% to 0%.[128] Differences in the frequency of the antigen in populations of Indigenous people in the Americas correlate with major language families, modified by environmental conditions.[129]
See also
- Introduction to genetics
- Archaeogenetics
- Archaeology of the Americas
- Ancient DNA
- Clovis culture
- Early human migrations
- Genetic history of Africa
- Genetic history of Europe
- Genetic history of Italy
- Genetic history of North Africa
- Genetic history of the British Isles
- Genetic history of the Iberian Peninsula
- Genetic history of the Middle East
- Genetics and archaeogenetics of South Asia
- List of haplogroups of historic people
- Race and genetics
- Settlement of the Americas § Genomic age estimates
- List of Y-chromosome haplogroups in populations of the world
Notes
- Algonquian ethnic groups: Ojibwe, Cheyenne/Arapaho, Shawnee, Mi'kmaq, Kickapoo and Meskwaki.
- Q-M3=12.9; Q(xM3)=20.6.
- Athabaskan ethnic groups: Chipewyan, Tłı̨chǫ, Tanana, Apache and Navajo.
- Q-M3=32.; Q3(xM3)=17.7.
- Chibchan ethnic groups: Ngöbe and Kuna peoples.
- Q-M3=6; Q(xM3)=25.
- P1(xQ) 62.5%. While other studies identify this as R(xR2)/R1b,
the subject remains controversial (see Hammer, Michael F. et al 2005) - Q-M3=8.2; Q(xQ-M3)=7.2.
- Q-M3=40.5; Q(xM3)=5.4.
- Gê ethnic groups: Gorotire, Kaigang, Kraho, Mekranoti and Xikrin.
- Q-M3=90; Q(xM3)=2)
- Q-M3=79; Q(xM3)=7.
- Q-M3=11; Q(xM3)=67.
- Q-M3=10.7; NWT01=44.6.
- Muskogean ethnic groups: Chickasaw, Choctaw, Muscogee and Seminole.
- Q-M3=50.0; Q(xM3)=25.0.
- Q-M3=83; Q(xM3)=9.
- Q-M3=89; Q(xM3)=11.
- C3*=9; C3b=9
- Q-M3=64; Q-MEH2*=9; Q-NWT01=9.
- Tupi–Guarani Brazilian ethnic groups: Asuriní, Parakanã, Ka'apor and Wayampi.
- All examples of haplogroup Q were Q-M3.
- Uto-Aztecan ethnic groups: Pima, Tohono O'odham, Tarahumara, Nahua, Cora and Huichol.
- Q=M3
- Q-M3=48; Q(xM3)=21.
- Q-M3=86<; Q(xM3)=14.
- Q=M3
- Q-M3=33; Q(xM3)=48.
References
- Wendy Tymchuk (2008). "Learn about Y-DNA Haplogroup Q. Genebase Tutorials". Genebase Systems. Archived from the original on June 22, 2010. Retrieved November 21, 2009.
- Orgel, Leslie E. (2004). "Prebiotic chemistry and the origin of the RNA world". Critical Reviews in Biochemistry and Molecular Biology. 39 (2): 99–123. CiteSeerX 10.1.1.537.7679. doi:10.1080/10409230490460765. PMID 15217990. S2CID 4939632.
- Tallbear, Kim (2014). "The Emergence, Politics, and Marketplace of Native American DNA". In Kleinman, Daniel Lee; Moore, Kelly (eds.). Routledge Handbook of Science, Technology, and Society. Routledge. p. 23. ISBN 978-1-136-23716-4. Archived from the original on January 30, 2016. Retrieved January 5, 2016.
- Moreno-Mayar, J. Víctor; Potter, Ben A.; Vinner, Lasse; et al. (January 2018). "Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans" (PDF). Nature. 553 (7687): 203–207. Bibcode:2018Natur.553..203M. doi:10.1038/nature25173. PMID 29323294. S2CID 4454580.
- Arnaiz-Villena, A.; Parga-Lozano, C.; Moreno, E.; Areces, C.; Rey, D.; Gomez-Prieto, P. (April 1, 2010). "The Origin of Amerindians and the Peopling of the Americas According to HLA Genes: Admixture with Asian and Pacific People". Current Genomics. Bentham Science Publishers Ltd. 11 (2): 103–114. doi:10.2174/138920210790886862. ISSN 1389-2029. PMC 2874220. PMID 20885818.
- Tamm, Erika; Kivisild, Toomas; Reidla, Maere; et al. (September 5, 2007). "Beringian Standstill and Spread of Native American Founders". PLOS ONE. 2 (9): e829. Bibcode:2007PLoSO...2..829T. doi:10.1371/journal.pone.0000829. PMC 1952074. PMID 17786201.
- Derenko, Miroslava; Malyarchuk, Boris; Grzybowski, Tomasz; et al. (December 21, 2010). "Origin and Post-Glacial Dispersal of Mitochondrial DNA Haplogroups C and D in Northern Asia". PLOS ONE. 5 (12): e15214. Bibcode:2010PLoSO...515214D. doi:10.1371/journal.pone.0015214. PMC 3006427. PMID 21203537.
- Bortolini, Maria-Catira; Salzano, Francisco M.; Thomas, Mark G.; et al. (September 2003). "Y-chromosome evidence for differing ancient demographic histories in the Americas". American Journal of Human Genetics. 73 (3): 524–539. doi:10.1086/377588. PMC 1180678. PMID 12900798.
- Ruhlen, Merritt (November 1998). "The Origin of the Na-Dene". Proceedings of the National Academy of Sciences of the United States of America. 95 (23): 13994–13996. Bibcode:1998PNAS...9513994R. doi:10.1073/pnas.95.23.13994. PMC 25007. PMID 9811914.
- Zegura, Stephen L.; Karafet, Tatiana M.; Zhivotovsky, Lev A.; et al. (January 2004). "High-Resolution SNPs and Microsatellite Haplotypes Point to a Single, Recent Entry of Native American Y Chromosomes into the Americas". Molecular Biology and Evolution. 21 (1): 164–175. doi:10.1093/molbev/msh009. PMID 14595095.
- Saillard, Juliette; Forster, Peter; Lynnerup, Niels; et al. (2000). "mtDNA Variation among Greenland Eskimos. The Edge of the Beringian Expansion". American Journal of Human Genetics. 67 (3): 718–726. doi:10.1086/303038. PMC 1287530. PMID 10924403.
- Schurr, Theodore G. (October 21, 2004). "The Peopling of the New World: Perspectives from Molecular Anthropology". Annual Review of Anthropology. 33: 551–583. doi:10.1146/annurev.anthro.33.070203.143932.
- Torroni, Antonio; Schurr, Theodore G.; Yang, Chi-Chuan; et al. (January 1992). "Native American Mitochondrial DNA Analysis Indicates That the Amerind and the Nadene Populations Were Founded by Two Independent Migrations". Genetics. 30 (1): 153–162. doi:10.1093/genetics/130.1.153. PMC 1204788. PMID 1346260.
- Wade, Nicholas (March 12, 2014). "Pause Is Seen in a Continent's Peopling". The New York Times. Archived from the original on April 9, 2021. Retrieved February 24, 2017.
- Lyovin, Anatole V. (1997). "Native Languages of the Americas". An Introduction to the Languages of the World. Oxford University. p. 309. ISBN 978-0-19-508115-2. Archived from the original on May 10, 2021. Retrieved June 7, 2020.
- Mithun, Marianne (October 1990). "Studies of North American Indian Languages". Annual Review of Anthropology. 19 (1): 309–330. doi:10.1146/annurev.an.19.100190.001521. JSTOR 2155968.
- Alice Roberts (2010). The Incredible Human Journey. A&C Black. pp. 101–03. ISBN 978-1-4088-1091-0. Archived from the original on January 25, 2021. Retrieved August 5, 2019.
- Wang, Sijia; Lewis, Cecil M. Jr.; Jakobsson, Mattias; et al. (November 23, 2007). "Genetic Variation and Population Structure in Native Americans". PLOS Genetics. 3 (11): e185. doi:10.1371/journal.pgen.0030185. PMC 2082466. PMID 18039031.
- Walsh, Bruce; Redd, Alan J.; Hammer, Michael F. (January 2008). "Joint match probabilities for Y chromosomal and autosomal markers". Forensic Science International. 174 (2–3): 234–238. doi:10.1016/j.forsciint.2007.03.014. PMID 17449208.
- Wells, Spencer (2002). The Journey of Man: A Genetic Odyssey. Princeton University Press. pp. 138–140. ISBN 978-0-691-11532-0.
- Hey, Jody (2005). "On the Number of New World Founders: A Population Genetic Portrait of the Peopling of the Americas". PLOS Biology. 3 (6): e193. doi:10.1371/journal.pbio.0030193. PMC 1131883. PMID 15898833.
- Wade, Nicholas (February 11, 2010). "Ancient Man In Greenland Has Genome Decoded". The New York Times. Archived from the original on March 8, 2021. Retrieved February 24, 2017.
- Reich, David; Patterson, Nick; Campbell, Desmond; et al. (August 16, 2012). "Reconstructing Native American population history". Nature. 488 (7411): 370–374. Bibcode:2012Natur.488..370R. doi:10.1038/nature11258. PMC 3615710. PMID 22801491.
- Rasmussen, Morten; Anzick, Sarah L.; Waters, Michael R.; et al. (2014). "The genome of a Late Pleistocene human from a Clovis burial site in western Montana". Nature. 506 (7487): 225–229. Bibcode:2014Natur.506..225R. doi:10.1038/nature13025. PMC 4878442. PMID 24522598.
- "Ancient American's genome mapped". BBC News. February 14, 2014. Archived from the original on May 5, 2021. Retrieved June 20, 2018.
- Dediu, Dan; Levinson, Stephen C. (September 20, 2012). "Abstract Profiles of Structural Stability Point to Universal Tendencies, Family-Specific Factors, and Ancient Connections between Languages". PLOS ONE. 7 (9): e45198. Bibcode:2012PLoSO...745198D. doi:10.1371/journal.pone.0045198. PMC 3447929. PMID 23028843.
- Raghavan, Maanasa; Steinrücken, Matthias; Harris, Kelley; et al. (August 21, 2015). "Genomic evidence for the Pleistocene and recent population history of Native Americans". Science. 349 (6250): aab3884. doi:10.1126/science.aab3884. PMC 4733658. PMID 26198033.
- Skoglund, Pontus; Mallick, Swapan; Bortolini, Maria Cátira; Chennagiri, Niru; Hünemeier, Tábita; Petzl-Erler, Maria Luiza; Salzano, Francisco Mauro; Patterson, Nick; Reich, David (September 2015). "Genetic evidence for two founding populations of the Americas". Nature. 525 (7567): 104–108. Bibcode:2015Natur.525..104S. doi:10.1038/nature14895. PMC 4982469. PMID 26196601.
- Skoglund, Pontus; Reich, David (December 2016). "A genomic view of the peopling of the Americas". Current Opinion in Genetics & Development. 41: 27–35. doi:10.1016/j.gde.2016.06.016. PMC 5161672. PMID 27507099.
- Gakuhari, Takashi; Nakagome, Shigeki; Rasmussen, Simon; et al. (December 2020). "Ancient Jomon genome sequence analysis sheds light on migration patterns of early East Asian populations". Communications Biology. 3 (1): 437. doi:10.1038/s42003-020-01162-2. PMC 7447786. PMID 32843717.
- Davis, Loren G.; Madsen, David B.; Becerra-Valdivia, Lorena; et al. (August 30, 2019). "Late Upper Paleolithic occupation at Cooper's Ferry, Idaho, USA, ~16,000 years ago". Science. 365 (6456): 891–897. Bibcode:2019Sci...365..891D. doi:10.1126/science.aax9830. PMID 31467216.
- Posth, Cosimo; Nakatsuka, Nathan; Lazaridis, Iosif; et al. (November 2018). "Reconstructing the Deep Population History of Central and South America". Cell. 175 (5): 1185–1197.e22. doi:10.1016/j.cell.2018.10.027. PMC 6327247. PMID 30415837.
- Willerslev, Eske; Meltzer, David J. (June 17, 2021). "Peopling of the Americas as inferred from ancient genomics". Nature. 594 (7863): 356–364. Bibcode:2021Natur.594..356W. doi:10.1038/s41586-021-03499-y. PMID 34135521. S2CID 235460793.
- Sarkar, Anjali A. (June 18, 2021). "Ancient Human Genomes Reveal Peopling of the Americas". GEN - Genetic Engineering and Biotechnology News. Retrieved September 15, 2021.
The team discovered that the Spirit Cave remains came from a Native American while dismissing a longstanding theory that a group called Paleoamericans existed in North America before Native Americans.
- Sapiens (February 8, 2022). "A Genetic Chronicle of the First Peoples in the Americas". SAPIENS. Retrieved April 3, 2023.
AROUND 36,000 years ago, a small group of people living in East Asia began to break off from the larger ancestral populations in the region. By about 25,000 years ago, the smaller group in East Asia itself split into two. One gave rise to a group referred to by geneticists as the ancient Paleo-Siberians, who stayed in Northeast Asia. The other became ancestral to Indigenous peoples in the Americas.
- Santos, Fabrício R.; Pandya, Arpita; Tyler-Smith, Chris; et al. (February 1999). "The Central Siberian Origin for Native American Y Chromosomes". American Journal of Human Genetics. 64 (2): 619–628. doi:10.1086/302242. PMC 1377773. PMID 9973301.
- Blanco Verea; Alejandro José. Linajes del cromosoma Y humano: aplicaciones genético-poblacionales y forenses. Univ Santiago de Compostela. pp. 135–. GGKEY:JCW0ASCR364. Archived from the original on January 30, 2016. Retrieved June 15, 2011.
- Zegura, Stephen L.; Karafet, Tatiana M.; Zhivotovsky, Lev A.; et al. (January 2004). "High-resolution SNPs and microsatellite haplotypes point to a single, recent entry of Native American Y chromosomes into the Americas". Molecular Biology and Evolution. 21 (1): 164–175. doi:10.1093/molbev/msh009. PMID 14595095.
- Saillard, Juliette; Forster, Peter; Lynnerup, Niels; et al. (September 2000). "mtDNA Variation among Greenland Eskimos: The Edge of the Beringian Expansion". The American Journal of Human Genetics. 67 (3): 718–726. doi:10.1086/303038. PMC 1287530. PMID 10924403.
- Schurr, Theodore G. (2004). "The Peopling of the New World – Perspectives from Molecular Anthropology". Annual Review of Anthropology. 33: 551–583. doi:10.1146/annurev.anthro.33.070203.143932. JSTOR 25064865. S2CID 4647888.
- Torroni, Antonio; Schurr, Theodore G.; Yang, Chi-Chuan; et al. (January 1992). "Native American Mitochondrial DNA Analysis Indicates That the Amerind and the Nadene Populations Were Founded by Two Independent Migrations". Genetics. 130 (1): 153–162. doi:10.1093/genetics/130.1.153. PMC 1204788. PMID 1346260.
- Balanovsky, Oleg; Gurianov, Vladimir; Zaporozhchenko, Valery; et al. (February 2017). "Phylogeography of human Y-chromosome haplogroup Q3-L275 from an academic/citizen science collaboration". BMC Evolutionary Biology. 17 (S1): 18. doi:10.1186/s12862-016-0870-2. PMC 5333174. PMID 28251872.
- "How the Subclades of Y-DNA Haplogroup Q are determined". Genebase Systems. 2009. Archived from the original on January 21, 2010. Retrieved November 22, 2009.
- "Y-DNA Haplogroup Tree". Genebase Systems. 2009. Archived from the original on December 25, 2010. Retrieved November 21, 2009.
- "Frequency Distribution of Y-DNA Haplogroup Q1a3a-M3". GeneTree. 2010. Archived from the original on November 4, 2009. Retrieved January 30, 2010.
- Flegontov, Pavel; Changmai, Piya; Zidkova, Anastassiya; Logacheva, Maria D.; Altınışık, N. Ezgi; Flegontova, Olga; Gelfand, Mikhail S.; Gerasimov, Evgeny S.; Khrameeva, Ekaterina E.; Konovalova, Olga P.; Neretina, Tatiana; Nikolsky, Yuri V.; Starostin, George; Stepanova, Vita V.; Travinsky, Igor V.; Tříska, Martin; Tříska, Petr; Tatarinova, Tatiana V. (2016). "Genomic study of the Ket: A Paleo-Eskimo-related ethnic group with significant ancient North Eurasian ancestry". Scientific Reports. 6: 20768. arXiv:1508.03097. Bibcode:2016NatSR...620768F. doi:10.1038/srep20768. PMC 4750364. PMID 26865217.
- "Learning Center :: Genebase Tutorials". Genebase.com. October 22, 1964. Archived from the original on November 17, 2013.
- Bonatto, SL; Salzano, FM (March 4, 1997). "A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data". Proceedings of the National Academy of Sciences of the United States of America. 94 (5): 1866–1871. Bibcode:1997PNAS...94.1866B. doi:10.1073/pnas.94.5.1866. PMC 20009. PMID 9050871.
- Smolenyak, Megan; Turner, Ann (2004). Trace Your Roots with DNA: Using Genetic Tests to Explore Your Family Tree. Rodale. p. 83. ISBN 978-1-59486-006-5. Archived from the original on January 30, 2016. Retrieved January 5, 2016.
- Than, Ker (February 14, 2008). "New World Settlers Took 20,000-Year Pit Stop". National Geographic News. Archived from the original on August 26, 2014. Retrieved January 24, 2010.
- Lovgren, Stefan (February 2, 2007). "First Americans Arrived Recently, Settled Pacific Coast, DNA Study Says". National Geographic News. Archived from the original on August 22, 2009. Retrieved February 2, 2010.
- "First Americans Endured 20,000-Year Layover – Jennifer Viegas, Discovery News". May 10, 2017. Archived from the original on October 10, 2012. Retrieved November 18, 2009. page 2 Archived March 13, 2012, at the Wayback Machine
- González Burchard, Esteban; Borrell, Luisa N.; Choudhry, Shweta; et al. (December 2005). "Latino Populations: A Unique Opportunity for the Study of Race, Genetics, and Social Environment in Epidemiological Research". American Journal of Public Health. 95 (12): 2161–2168. doi:10.2105/AJPH.2005.068668. PMC 1449501. PMID 16257940.
- Malhi, Ripan Singh; Gonzalez-Oliver, Angelica; Schroeder, Kari Britt; et al. (December 2008). "Distribution of Y chromosomes among Native North Americans: A study of Athapaskan population history". American Journal of Physical Anthropology. 137 (4): 412–424. doi:10.1002/ajpa.20883. PMC 2584155. PMID 18618732.
- Bolnick, Deborah; Bolnick, Daniel; Smith, David (2006). "Asymmetric Male and Female Genetic Histories among Native Americans from Eastern North America". Molecular Biology and Evolution. 23 (11): 2161–2174. doi:10.1093/molbev/msl088. PMID 16916941. "In most cases, there is widespread agreement about whether a particular haplogroup represents an ancient Native American lineage or post-1492 admixture, but the status of haplogroup R-M173 has recently been subject to some debate. Some authors have argued that this haplogroup represents a founding Native American lineage (Lell et al. 2002; Bortolini et al. 2003), whereas others suggest that it instead reflects recent European admixture (Tarazona-Santos and Santos 2002; Bosch et al. 2003; Zegura et al. 2004). In eastern North America, the pattern of haplotype variation within this haplogroup supports the latter hypothesis: R-M173 haplotypes do not cluster by population or culture area, as haplotypes in the other founding haplogroups do, and most match or are closely related to R-M173 haplotypes that are common in Europe but rare in Asia. This pattern is opposite than expected if the Native American R-M173 haplotypes were descended from Asian haplotypes and suggests that recent European admixture is responsible for the presence of haplogroup R-M173 in eastern North America."
- Bolnick, Deborah Ann (2005). The Genetic Prehistory of Eastern North America: Evidence from Ancient and Modern DNA. University of California, Davis. p. 83. "Haplogroup R - M173 likely represents recent (post 1492) European admixture, as may P-M45* (Tarazona-Santos and Santos 2002; Bosch et al. 2003; Zegura et al. 2004)"
- Raff 2022, pp. 59–60:"Y chromosome founder haplogroups in Native Americans include Q-M3 (and its sub-haplogroups, Q-CTS1780), and C3-MPB373 (potentially C- P39-Z30536). Other haplogroups found [sic] Native American populations, like R1b, were likely the result of post-European contact admixture (44)."
- Malhi et al. 2008: "All individuals that did not belong to haplogroup Q and C were excluded from the Haplotype data set because these haplotypes are likely the result of non-native admixture (Tarazona-Santos and Santos, 2002; Zegura et al., 2004; Bolnick et al, 2006)...The frequency of haplogroup C is highest in Northwestern North America and the frequency of haplogroup R, the presence of which is attributed to European admixture, reaches its maximum in Northeastern North America."
- Zegura, S. L. (October 31, 2003). "High-Resolution SNPs and Microsatellite Haplotypes Point to a Single, Recent Entry of Native American Y Chromosomes into the Americas". Molecular Biology and Evolution. Oxford University Press (OUP). 21 (1): 164–175. doi:10.1093/molbev/msh009. ISSN 0737-4038. PMID 14595095.
- Zhong, Hua; Shi, Hong; Qi, Xue-Bin; et al. (July 2010). "Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia". Journal of Human Genetics. 55 (7): 428–435. doi:10.1038/jhg.2010.40. PMID 20448651. S2CID 28609578.
- Xue, Yali; Zerjal, Tatiana; Bao, Weidong; et al. (April 1, 2006). "Male Demography in East Asia: a North-South Contrast in Human Population Expansion Times". Genetics. 172 (4): 2431–2439. doi:10.1534/genetics.105.054270. PMC 1456369. PMID 16489223.
- Marinakis, Aliki (2008). "Na-Dene People". In Johansen, Bruce E.; Pritzker, Barry M. (eds.). Encyclopedia of American Indian History. ABC-CLIO. p. 441. ISBN 978-1-85109-818-7. Archived from the original on January 30, 2016. Retrieved January 5, 2016.
- ISOGG, 2015 "Y-DNA Haplogroup C and its Subclades – 2015" Archived 2021-08-15 at the Wayback Machine (15 September 2015).
- Zegura, S. L. (October 31, 2003). "High-Resolution SNPs and Microsatellite Haplotypes Point to a Single, Recent Entry of Native American Y Chromosomes into the Americas". Molecular Biology and Evolution. 21 (1): 164–175. doi:10.1093/molbev/msh009. PMID 14595095.
- Bolnick, D. A. (August 10, 2006). "Asymmetric Male and Female Genetic Histories among Native Americans from Eastern North America". Molecular Biology and Evolution. 23 (11): 2161–2174. doi:10.1093/molbev/msl088. PMID 16916941. S2CID 30220691.
- Dulik, M. C.; Owings, A. C.; Gaieski, J. B.; et al. (May 14, 2012). "Y-chromosome analysis reveals genetic divergence and new founding native lineages in Athapaskan- and Eskimoan-speaking populations". Proceedings of the National Academy of Sciences. 109 (22): 8471–8476. Bibcode:2012PNAS..109.8471D. doi:10.1073/pnas.1118760109. PMC 3365193. PMID 22586127.
- Hammer, Michael F.; Chamberlain, Veronica F.; Kearney, Veronica F.; et al. (December 2006). "Population structure of Y chromosome SNP haplogroups in the United States and forensic implications for constructing Y chromosome STR databases". Forensic Science International. 164 (1): 45–55. doi:10.1016/j.forsciint.2005.11.013. PMID 16337103.
- Schurr, Theodore G. (May–June 2000). "Mitochondrial DNA and the Peopling of the New World". American Scientist. 88 (3): 246. doi:10.1511/2000.23.772.
- Zakharov, Ilia A.; Derenko, Miroslava V.; Maliarchuk, Boris A.; et al. (April 2004). "Mitochondrial DNA variation in the aboriginal populations of the Altai-Baikal region: implications for the genetic history of North Asia and America". Annals of the New York Academy of Sciences. 1011 (1): 21–35. Bibcode:2004NYASA1011...21Z. doi:10.1196/annals.1293.003. PMID 15126280. S2CID 37139929.
- Starikovskaya, Elena B.; Sukernik, Rem I.; Derbeneva, Olga A.; et al. (January 7, 2005). "Mitochondrial DNA diversity in Indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups". Annals of Human Genetics. 69 (1): 67–89. doi:10.1046/j.1529-8817.2003.00127.x. PMC 3905771. PMID 15638829.
- Felton, James (May 9, 2023). "DNA Sheds Light On Mystery About Where Native Americans Came From". IFLScience. Retrieved May 24, 2023.
- Eshleman, Jason A.; Malhi, Ripan S.; Smith, David Glenn (February 14, 2003). "Mitochondrial DNA Studies of Native Americans- Conceptions and Misconceptions of the Population Prehistory of the Americas". Evolutionary Anthropology. 12 (1): 7–18. doi:10.1002/evan.10048. S2CID 17049337.
- Achilli, Alessandro; Perego, Ugo A.; Bravi, Claudio M.; et al. (March 12, 2008). "The Phylogeny of the Four Pan-American MtDNA Haplogroups: Implications for Evolutionary and Disease Studies". PLOS ONE. 3 (3): e1764. Bibcode:2008PLoSO...3.1764A. doi:10.1371/journal.pone.0001764. PMC 2258150. PMID 18335039.
- Nina G. Jablonski (2002). The first Americans: the Pleistocene colonization of the New World. University of California Press. p. 301. ISBN 978-0-940228-50-4. Archived from the original on January 30, 2016. Retrieved June 15, 2011.
- Scatena, Roberto; Bottoni, Patrizia; Giardina, Bruno (March 8, 2012). Advances in Mitochondrial Medicine. Springer Science & Business Media. p. 446. ISBN 978-94-007-2869-1.
- Raff, Jennifer A.; Bolnick, Deborah A. (October 2015). "Does Mitochondrial Haplogroup X Indicate Ancient Trans-Atlantic Migration to the Americas? A Critical Re-Evaluation". PaleoAmerica. 1 (4): 297–304. doi:10.1179/2055556315Z.00000000040. ISSN 2055-5563. S2CID 85626735. "These studies have all reached the same conclusion and suggest that haplogroup X2a is likely to have originated in the same population(s) as the other American founder haplogroups, by virtue of having comparable coalescence dates and demographic histories" ... "X2a has not been found anywhere in Eurasia, and phylogeography gives us no compelling reason to think it is more likely to come from Europe than from Siberia. Furthermore, analysis of the complete genome of Kennewick Man, who belongs to the most basal lineage of X2a yet identified, gives no indication of recent European ancestry and moves the location of the deepest branch of X2a to the West Coast, consistent with X2a belonging to the same ancestral population as the other founder mitochondrial haplogroups."
- "The peopling of the Americas: Genetic ancestry influences health". Scientific American. August 14, 2009. Archived from the original on September 14, 2011. Retrieved January 19, 2010 – via Phys.org.
- Fagundes, Nelson J.R.; Kanitz, Ricardo; Eckert, Roberta; et al. (March 3, 2008). "Mitochondrial Population Genomics Supports a Single Pre-Clovis Origin with a Coastal Route for the Peopling of the Americas". American Journal of Human Genetics. 82 (3): 583–592. doi:10.1016/j.ajhg.2007.11.013. PMC 2427228. PMID 18313026.
- Meltzer, David J. (2009). First Peoples in a New World: Colonizing Ice Age America. University of California Press. p. 162. ISBN 978-0-520-94315-5. Archived from the original on January 30, 2016. Retrieved January 5, 2016.
- "An mtDNA view of the peopling of the world by Homo sapiens". Cambridge DNA. 2009. Archived from the original on May 11, 2011. Retrieved April 29, 2010.
- Reidla, Maere; Kivisild, Toomas; Metspalu, Ene; et al. (November 2003). "Origin and Diffusion of mtDNA Haplogroup X". American Journal of Human Genetics. 73 (5): 1178–1190. doi:10.1086/379380. PMC 1180497. PMID 14574647.
- "An mtDNA view of the peopling of the world by Homo sapiens". Cambridge DNA Services. 2007. Archived from the original on May 11, 2011. Retrieved June 1, 2011.
- Peñaloza-Espinosa, Rosenda I.; Arenas-Aranda, Diego; Cerda-Flores, Ricardo M.; et al. (2007). "Characterization of mtDNA Haplogroups in 14 Mexican Indigenous Populations". Human Biology. 79 (3): 313–320. doi:10.1353/hub.2007.0042. PMID 18078204. S2CID 35654242.
- Ferrell, Robert E.; Chakraborty, Ranajit; Gershowitz, Henry; et al. (July 1981). "The St. Lawrence Island Eskimos: Genetic variation and genetic distance". American Journal of Physical Anthropology. 55 (3): 351–358. doi:10.1002/ajpa.1330550309. PMID 6455922.
- Helgason, Agnar; Pálsson, Gísli; Pedersen, Henning Sloth; et al. (May 2006). "mtDNA variation in Inuit populations of Greenland and Canada: Migration history and population structure". American Journal of Physical Anthropology. 130 (1): 123–134. doi:10.1002/ajpa.20313. PMID 16353217.
- Raghavan, Maanasa; Skoglund, Pontus; Graf, Kelly E.; et al. (January 2014). "Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans". Nature. 505 (7481): 87–91. Bibcode:2014Natur.505...87R. doi:10.1038/nature12736. PMC 4105016. PMID 24256729.
- Kashani, Baharak Hooshiar; Perego, Ugo A.; Olivieri, Anna; et al. (January 2012). "Mitochondrial haplogroup C4c: A rare lineage entering America through the ice-free corridor?". American Journal of Physical Anthropology. 147 (1): 35–39. doi:10.1002/ajpa.21614. PMID 22024980.
- Llamas, Bastien; Fehren-Schmitz, Lars; Valverde, Guido; et al. (April 29, 2016). "Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas". Science Advances. 2 (4): e1501385. Bibcode:2016SciA....2E1385L. doi:10.1126/sciadv.1501385. PMC 4820370. PMID 27051878.
- "Direct genetic evidence of founding population reveals story of first Native Americans". University of Cambridge. January 3, 2018. Archived from the original on September 25, 2018. Retrieved October 15, 2018.
- "Direct genetic evidence of founding population reveals story of first Native Americans". Archived from the original on November 5, 2018. Retrieved October 15, 2018.
- García-Bour, Jaume; Pérez-Pérez, Alejandro; Álvarez, Sara; et al. (2004). "Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: Ancient mtDNA sequences and Y-Chromosome STR characterization". American Journal of Physical Anthropology. 123 (4): 361–370. doi:10.1002/ajpa.10337. PMID 15022364.
- Neves, W.A.; Powell, J.F.; Ozolins, E.G. (1999). "Extra-continental morphological affinities of Palli Aike, southern Chile". Interciencia. 24 (4): 258–263. Archived from the original on July 9, 2021. Retrieved July 8, 2021.
- Bonatto, Sandro L.; Salzano, Francisco M. (March 4, 1997). "A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data". Proceedings of the National Academy of Sciences. 94 (5): 1866–1871. Bibcode:1997PNAS...94.1866B. doi:10.1073/pnas.94.5.1866. PMC 20009. PMID 9050871.
- Cinq-Mars, J. (1979). "Bluefish Cave I: A Late Pleistocene Eastern Beringian Cave Deposit in the Northern Yukon". Canadian Journal of Archaeology (3): 1–32. JSTOR 41102194.
- Bonnichsen, Robson (1978). "Critical arguments for Pleistocene artifacts from the Old Crow basin, Yukon: a preliminary statement". In Bryan, Alan Lyle (ed.). Early Man in America from a Circum-pacific Perspective. Archaeological Researches International. pp. 102–118. ISBN 978-0-88864-999-7.
- Dillehay, Tom D.; Ocampo, Carlos (2015). "New Archaeological Evidence for an Early Human Presence at Monte Verde, Chile". PLOS ONE. 10 (11): e0141923. Bibcode:2015PLoSO..1041923D. doi:10.1371/journal.pone.0141923. PMC 4651426. PMID 26580202.
- Bourgeon, Lauriane; Burke, Ariane; Higham, Thomas (January 6, 2017). "Earliest Human Presence in North America Dated to the Last Glacial Maximum: New Radiocarbon Dates from Bluefish Caves, Canada". PLOS ONE. 12 (1): e0169486. Bibcode:2017PLoSO..1269486B. doi:10.1371/journal.pone.0169486. PMC 5218561. PMID 28060931.
- Gilbert, M. Thomas P.; Jenkins, Dennis L.; Götherstrom, Anders; et al. (May 9, 2008). "DNA from Pre-Clovis Human Coprolites in Oregon, North America". Science. 320 (5877): 786–789. Bibcode:2008Sci...320..786G. doi:10.1126/science.1154116. PMID 18388261. S2CID 17671309.
- Newman, Richard (1999). "Miscegenation". In Kwame Appiah and Henry Louis Gates, Jr. (ed.). Africana: The Encyclopedia of the African and African American Experience (1st ed.). New York: Basic Civitas Books. p. 1320. ISBN 978-0-465-00071-5.
Miscegenation, a term for sexual relations across racial lines; no longer in use because of its racist implications
- Chasteen, John Charles; Wood, James A (2004). Problems in modern Latin American history, sources and interpretations. Sr Books. pp. 4–10. ISBN 978-0-8420-5060-9. Archived from the original on August 15, 2021. Retrieved February 24, 2010.
- Chasteen (2004:4): "between the White elite and the mass of [Indigenous Americans] and Negroes there existed by 1700 a thin stratum of population subject neither to Negro slavery nor [Indigenous American] tutelage, consisting of the products of racial interbreeding among Whites, [Indigenous Americans], and Negroes and defined as mestizos, mulattoes and zambos (mixture of [Indigenous American] and Negro) and their many combinations."
- Sans, Mónica (2000). "Admixture Studies in Latin America: From the 20th to the 21st Century". Human Biology. 72 (1): 155–177. JSTOR 41465813. PMID 10721616.
- Sweet, Frank W (2004). "Afro-European Genetic Admixture in the United States". Essays on the Color Line and the One-Drop Rule. Backintyme Essays. Archived from the original on January 4, 2010. Retrieved February 24, 2010.
- Sweet, Frank W (2005). Legal History of the Color Line: The Notion of Invisible Blackness. Backintyme Publishing. p. 542. ISBN 978-0-939479-23-8. Archived from the original on February 17, 2010. Retrieved February 24, 2010.
- Soong, Roland (1999). "Racial Classifications in Latin America". Latin American Media. Archived from the original on April 30, 2010. Retrieved February 25, 2010.
- Brehaut, Harry B (1998). "The Red River Cart and Trails: The Fur Trade". Manitoba Historical Society. Archived from the original on July 9, 2011. Retrieved February 24, 2010.
- "Who are Métis ?". Métis National Council. 2001. Archived from the original on February 26, 2010. Retrieved February 24, 2010.
- Kimberly TallBear (2003). "DNA, Blood, and Racializing the Tribe". Wíčazo Ša Review. University of Minnesota Press. 18 (1): 81–107. doi:10.1353/wic.2003.0008. JSTOR 140943. S2CID 201778441.
- Furukawa, Julia (May 22, 2023). "Review of genealogies, other records fails to support local leaders' claims of Abenaki ancestry". New Hampshire Public Radio. Retrieved July 7, 2023.
- Adams, Paul (July 10, 2011). "Blood quantum influences Native American identity". BBC News. Archived from the original on January 29, 2023. Retrieved October 22, 2017.
- Bureau, U.S. Census. "U.S. Census website". United States Census Bureau. Archived from the original on December 27, 1996. Retrieved October 22, 2017.
- "Why Do So Many People Claim They Have Cherokee In Their Blood? - Nerve". www.nerve.com. Archived from the original on October 22, 2017. Retrieved October 22, 2017.
- Smithers, Gregory D. (October 1, 2015). "Why Do So Many Americans Think They Have Cherokee Blood?". Slate. Archived from the original on October 5, 2018. Retrieved October 22, 2017.
- "The Cherokee Syndrome - Daily Yonder". www.dailyyonder.com. February 10, 2011. Archived from the original on October 21, 2017. Retrieved October 22, 2017.
- Hitt, Jack (August 21, 2005). "The Newest Indians". The New York Times. Archived from the original on October 21, 2017. Retrieved October 22, 2017 – via www.nytimes.com.
- Nieves, Evelyn (March 3, 2007). "Putting to a Vote the Question 'Who Is Cherokee?'". The New York Times. Archived from the original on August 30, 2017. Retrieved October 22, 2017 – via www.nytimes.com..
- "What Percentage Indian Do You Have to Be in Order to Be a Member of a Tribe or Nation? - Indian Country Media Network". indiancountrymedianetwork.com. Archived from the original on October 21, 2017. Retrieved October 22, 2017.. "Disappearing Indians, Part II: The Hypocrisy of Race In Deciding Who's Enrolled - Indian Country Media Network". indiancountrymedianetwork.com. Archived from the original on September 22, 2017. Retrieved October 22, 2017.
- Price, Michael (November 15, 2016). "European diseases left a genetic mark on Native Americans". Science. doi:10.1126/science.aal0382.
- Hershey A, Chase M (1952). "Independent functions of viral protein and nucleic acid in growth of bacteriophage". J Gen Physiol. 36 (1): 39–56. doi:10.1085/jgp.36.1.39. PMC 2147348. PMID 12981234.
- Lille-Szyszkowicz I (1957). "Rozwój badan nad plejadami grup krwi" [Development of studies on pleiades of blood groups]. Postępy Higieny i Medycyny Doświadczalnej (in Polish). 11 (3): 229–33. OCLC 101713985. PMID 13505351.
- Landsteiner, Karl (1900). "Zur Kenntnis der antifermentativen, lytischen und agglutinierenden Wirkungen des Blutserums und der Lymphe" [Knowledge of the antifermentative, lytic and agglutinating effects of blood serum and lymph]. Zentralblatt für Bakteriologie (in German). 27: 357–362. OCLC 11337636. NAID 10008616088.
- Yazer M, Olsson M, Palcic M (2006). "The cis-AB blood group phenotype: fundamental lessons in glycobiology". Transfus Med Rev. 20 (3): 207–17. doi:10.1016/j.tmrv.2006.03.002. PMID 16787828.
- "Distribution of Blood Types". Behavioral Sciences Department, Palomar College. 2010. Archived from the original on February 21, 2006. Retrieved March 14, 2010.
- Estrada-Mena B, Estrada FJ, Ulloa-Arvizu R, et al. (May 2010). "Blood group O alleles in Native Americans: implications in the peopling of the Americas". Am. J. Phys. Anthropol. 142 (1): 85–94. doi:10.1002/ajpa.21204. PMID 19862808.
- Chown, Bruce; Lewis, Marion (June 1956). "The blood group genes of the Cree Indians and the Eskimos of the ungava district of Canada". American Journal of Physical Anthropology. 14 (2): 215–224. doi:10.1002/ajpa.1330140217. PMID 13362488.
- Halverson, Melissa S.; Bolnick, Deborah A. (November 2008). "An ancient DNA test of a founder effect in Native American ABO blood group frequencies". American Journal of Physical Anthropology. 137 (3): 342–347. doi:10.1002/ajpa.20887. PMID 18618657.
- "Racial and Ethnic Distribution of ABO Blood Types". BloodBook. 2008. Archived from the original on March 4, 2010. Retrieved March 31, 2010.
- Poole J (2020). "The Diego blood group system-an update". Immunohematology. 15 (4): 135–43. doi:10.21307/immunohematology-2019-635. PMID 15373634.
- Bégat C, Bailly P, Chiaroni J, Mazières S (July 6, 2015). "Revisiting the Diego Blood Group System in Amerindians: Evidence for Gene-Culture Comigration". PLOS ONE. 10 (7): e0132211. Bibcode:2015PLoSO..1032211B. doi:10.1371/journal.pone.0132211. PMC 4493026. PMID 26148209.
Further reading
- Peter N. Jones (October 2002). American Indian mtDNA, Y chromosome genetic data, and the peopling of North America. Bauu Institute. ISBN 978-0-9721349-1-0.
- Joseph Frederick Powell (2005). The first Americans: race, evolution, and the origin of Native Americans. Cambridge University Press. ISBN 978-0-521-82350-0.
- Francisco M. Salzano; Maria Cátira Bortolini (2002). The evolution and genetics of Latin American populations. Cambridge University Press. ISBN 978-0-521-65275-9.
- "The peopling of the Americas: Genetic ancestry influences health". American Journal of Physical Anthropology. University of Oklahoma. 2009. Retrieved November 21, 2009.
- McInnes, Roderick R. (March 2011). "2010 Presidential Address: Culture: The Silent Language Geneticists Must Learn— Genetic Research with Indigenous Populations". The American Journal of Human Genetics. 88 (3): 254–261. doi:10.1016/j.ajhg.2011.02.014. PMC 3059421. PMID 21516613.
- Raff, Jennifer (February 8, 2022). Origin: A Genetic History of the Americas. Grand Central Publishing. ISBN 978-1-5387-4970-8.
.svg.png.webp)