Gibberellin
Gibberellins (GAs) are plant hormones that regulate various developmental processes, including stem elongation, germination, dormancy, flowering, flower development, and leaf and fruit senescence.[1] GAs are one of the longest-known classes of plant hormone. It is thought that the selective breeding (albeit unconscious) of crop strains that were deficient in GA synthesis was one of the key drivers of the "green revolution" in the 1960s,[2] a revolution that is credited to have saved over a billion lives worldwide.[3]
History
The first inroads into the understanding of GAs were developments from the plant pathology field, with studies on the bakanae, or "foolish seedling" disease in rice. Foolish seedling disease causes a strong elongation of rice stems and leaves and eventually causes them to topple over.[4] In 1926, Japanese scientist Eiichi Kurosawa identified that foolish seedling disease was caused by the fungus Gibberella fujikuroi.[4] Later work at the University of Tokyo showed that a substance produced by this fungus triggered the symptoms of foolish seedling disease and they named this substance "gibberellin".[1][4]
The increased communication between Japan and the West following World War II enhanced the interest in gibberellin in the United Kingdom (UK) and the United States (US).[1] Workers at Imperial Chemical Industries in the UK[5] and the Department of Agriculture in the US both independently isolated gibberellic acid[4] (with the Americans originally referring to the chemical as "gibberellin-X", before adopting the British name–the chemical is known as gibberellin A3 or GA3 in Japan)[1]
Knowledge of gibberellins spread around the world as the potential for its use on various commercially important plants became more obvious. For example, in 1957, a viticulture professor at the University of California, Davis reported on test results showing that gibberellin had uses in the production of grapes, particularly of Thompson seedless table grapes. Five years later, all Thompson seedless table grapes in California were being sprayed with gibberellin at fruit set to increase berry size.[6] A known gibberellin biosynthesis inhibitor is paclobutrazol (PBZ), which in turn inhibits growth and induces early fruitset as well as seedset.
A chronic food shortage was feared during the rapid climb in world population in the 1960s. This was averted with the development of a high-yielding variety of rice. This variety of semi-dwarf rice is called IR8, and it has a short height because of a mutation in the sd1 gene.[7] Sd1 encodes GA20ox, so a mutant sd1 is expected to exhibit a short height that is consistent with GA deficiency.[2]
Chemistry
All known gibberellins are diterpenoid acids that are synthesized by the terpenoid pathway in plastids and then modified in the endoplasmic reticulum and cytosol until they reach their biologically active form.[8] All gibberellins are derived via the ent-gibberellane skeleton, but are synthesised via ent-kaurene. The gibberellins are named GA1 through GAn in order of discovery.[9] Gibberellic acid, which was the first gibberellin to be structurally characterized, is GA3.[10]
As of 2020,[9] there are 136 GAs identified from plants, fungi, and bacteria.[1][10][9]
Gibberellins are tetracyclic diterpene acids. There are two classes based on the presence of either 19 or 20 carbons. The 19-carbon gibberellins, such as gibberellic acid, have lost carbon 20 and, in place, possess a five-member lactone bridge that links carbons 4 and 10. The 19-carbon forms are, in general, the biologically active forms of gibberellins. Hydroxylation also has a great effect on the biological activity of the gibberellin. In general, the most biologically active compounds are dihydroxylated gibberellins, which possess hydroxyl groups on both carbon 3 and carbon 13. Gibberellic acid is a dihydroxylated gibberellin.[11]
Bioactive GAs
The bioactive GAs are GA1, GA3, GA4, and GA7.[12] There are three common structural traits between these GAs: hydroxyl group on C-3β, a carboxyl group on C-6, and a lactone between C-4 and C-10.[12] The 3β-hydroxyl group can be exchanged for other functional groups at C-2 and/or C-3 positions.[12] GA5 and GA6 are examples of bioactive GAs that do not have a hydroxyl group on C-3β.[12] The presence of GA1 in various plant species suggests that it is a common bioactive GA.[13]
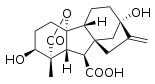 Gibberellin A1 (GA1)
Gibberellin A1 (GA1) Gibberellic acid (GA3)
Gibberellic acid (GA3)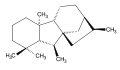 ent-Gibberellane
ent-Gibberellane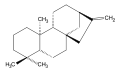 ent-Kaurene
ent-Kaurene
Biological function

Gibberellins are involved in the natural process of breaking dormancy and other aspects of germination. Before the photosynthetic apparatus develops sufficiently in the early stages of germination, the stored energy reserves of starch nourish the seedling. Usually in germination, the breakdown of starch to glucose in the endosperm begins shortly after the seed is exposed to water.[14] Gibberellins in the seed embryo are believed to signal starch hydrolysis through inducing the synthesis of the enzyme α-amylase in the aleurone cells. In the model for gibberellin-induced production of α-amylase, it is demonstrated that gibberellins (denoted by GA) produced in the scutellum diffuse to the aleurone cells, where they stimulate the secretion α-amylase.[8] α-Amylase then hydrolyses starch, which is abundant in many seeds, into glucose that can be used in cellular respiration to produce energy for the seed embryo. Studies of this process have indicated gibberellins cause higher levels of transcription of the gene coding for the α-amylase enzyme, to stimulate the synthesis of α-amylase.[11]
Gibberellins are produced in greater mass when the plant is exposed to cold temperatures. They stimulate cell elongation, breaking and budding, seedless fruits, and seed germination. Gibberellins cause seed germination by breaking the seed's dormancy and acting as a chemical messenger. Its hormone binds to a receptor, and calcium activates the protein calmodulin, and the complex binds to DNA, producing an enzyme to stimulate growth in the embryo.
Metabolism
Biosynthesis
GAs are usually synthesized from the methylerythritol phosphate (MEP) pathway in higher plants.[15] In this pathway, bioactive GA is produced from trans-geranylgeranyl diphosphate (GGDP).[15] In the MEP pathway, three classes of enzymes are used to yield GA from GGDP: terpene syntheses (TPSs), cytochrome P450 monooxygenases (P450s), and 2-oxoglutarate–dependent dioxygenases (2ODDs).[12] There are eight steps in the MEP pathway:[12]
- GGDP is converted to ent-copalyl diphosphate (ent-CDP) by ent-copalyl diphosphate synthase (CPS)
- ent-CDP is converted to ent-kaurene by ent-kaurene synthase (KS)
- ent-kaurene is converted to ent-kaurenol by ent-kaurene oxidase (KO)
- ent-kaurenol is converted to ent-kaurenal by KO
- ent-kaurenal is converted to ent-kaurenoic acid by KO
- ent-kaurenoic acid is converted to ent-7a-hydroxykaurenoic acid by ent-kaurenoic acid oxidase (KAO)
- ent-7a-hydroxykaurenoic acid is converted to GA12-aldehyde by KAO
- GA12-aldehyde is converted to GA12 by KAO. GA12 is processed to the bioactive GA4 by oxidations on C-20 and C-3, which is accomplished by 2 soluble ODDs: GA 20-oxidase and GA 3-oxidase.
One or two genes encode the enzymes responsible for the first steps of GA biosynthesis in Arabidopsis and rice.[12] The null alleles of the genes encoding CPS, KS, and KO result in GA-deficient Arabidopsis dwarves.[16] Multigene families encode the 2ODDs that catalyze the formation of GA12 to bioactive GA4.[12]
AtGA3ox1 and AtGA3ox2, two of the four genes that encode GA3ox in Arabidopsis, affect vegetative development.[17] Environmental stimuli regulate AtGA3ox1 and AtGA3ox2 activity during seed germination.[18][19] In Arabidopsis, GA20ox overexpression leads to an increase in GA concentration.[20][21]
Sites of biosynthesis
Most bioactive GAs are located in actively growing organs on plants.[15] Both GA20ox and GA3ox genes (genes coding for GA 20-oxidase and GA 3-oxidase) and the SLENDER1 gene (a GA signal transduction gene) are found in growing organs on rice, which suggests bioactive GA synthesis occurs at their site of action in growing organs in plants.[22] During flower development, the tapetum of anthers is believed to be a primary site of GA biosynthesis.[22][23]
Differences between biosynthesis in fungi and lower plants
Arabidopsis, a plant, and Gibberella fujikuroi, a fungus, possess different GA pathways and enzymes.[12] P450s in fungi perform functions analogous to the functions of KAOs in plants.[24] The function of CPS and KS in plants is performed by a single enzyme, CPS/KS, in fungi.[25][26][27] In fungi, the GA biosynthesis genes are found on one chromosome, but in plants, they are found randomly on multiple chromosomes.[28][29] Plants produce low amount of GA3, therefore the GA3 is produced for industrial purposes by microorganisms. Industrially the gibberellic acid can be produced by submerged fermentation, but this process presents low yield with high production costs and hence higher sale value, nevertheless other alternative process to reduce costs of the GA3 production is solid-state fermentation (SSF) that allows the use of agro-industrial residues.[30]
Catabolism
Several mechanisms for inactivating GAs have been identified. 2β-hydroxylation deactivates GA, and is catalyzed by GA2-oxidases (GA2oxs).[15] Some GA2oxs use C19-GAs as substrates, and other GA2oxs use C20-GAs.[31][32] Cytochrome P450 mono-oxygenase, encoded by elongated uppermost internode (eui), converts GAs into 16α,17-epoxides.[33] Rice eui mutants amass bioactive GAs at high levels, which suggests cytochrome P450 mono-oxygenase is a main enzyme responsible for deactivation GA in rice.[33] The Gamt1 and gamt2 genes encode enzymes that methylate the C-6 carboxyl group of GAs.[34] In a gamt1 and gamt2 mutant, concentrations of GA is developing seeds is increased.[34]
Homeostasis
Feedback and feedforward regulation maintains the levels of bioactive GAs in plants.[35][36] Levels of AtGA20ox1 and AtGA3ox1 expression are increased in a GA deficient environment, and decreased after the addition of bioactive GAs,[18][37][38][39][40] Conversely, expression of AtGA2ox1 and AtGA2ox2, GA deactivation genes, is increased with addition of GA.[31]
Regulation
Regulation by other hormones
The auxin indole-3-acetic acid (IAA) regulates concentration of GA1 in elongating internodes in peas.[41] Removal of IAA by removal of the apical bud, the auxin source, reduces the concentration of GA1, and reintroduction of IAA reverses these effects to increase the concentration of GA1.[41] This phenomenon has also been observed in tobacco plants.[42] Auxin increases GA 3-oxidation and decreases GA 2-oxidation in barley.[43] Auxin also regulates GA biosynthesis during fruit development in peas.[44] These discoveries in different plant species suggest the auxin regulation of GA metabolism may be a universal mechanism.
Ethylene decreases the concentration of bioactive GAs.[45]
Regulation by environmental factors
Recent evidence suggests fluctuations in GA concentration influence light-regulated seed germination, photomorphogenesis during de-etiolation, and photoperiod regulation of stem elongation and flowering.[12] Microarray analysis showed about one fourth cold-responsive genes are related to GA-regulated genes, which suggests GA influences response to cold temperatures.[19] Plants reduce growth rate when exposed to stress. A relationship between GA levels and amount of stress experienced has been suggested in barley.[46]
Role in seed development
Bioactive GAs and abscisic acid levels have an inverse relationship and regulate seed development and germination.[47][48] Levels of FUS3, an Arabidopsis transcription factor, are upregulated by ABA and downregulated by GA, which suggests that there is a regulation loop that establishes the balance of GA and ABA.[49]
Signalling mechanism
Receptor
In the early 1990s, there were several lines of evidence that suggested the existence of a GA receptor in oat seeds that was located at the plasma membrane. However, despite intensive research, to date, no membrane-bound GA receptor has been isolated. This, along with the discovery of a soluble receptor, GA insensitive dwarf 1 (GID1) has led many to doubt that a membrane-bound receptor exists.[1]

GID1 was first identified in rice[50] and in Arabidopsis there are three orthologs of GID1, AtGID1a, b, and c.[1] GID1s have a high affinity for bioactive GAs.[50] GA binds to a specific binding pocket on GID1; the C3-hydroxyl on GA makes contact with tyrosine-31 in the GID1 binding pocket.[51][52] GA binding to GID1 causes changes in GID1 structure, causing a 'lid' on GID1 to cover the GA binding pocket. The movement of this lid results in the exposure of a surface which enables the binding of GID1 to DELLA proteins.[51][52]
DELLA proteins: Repression of a repressor
DELLA proteins, such as SLR1 in rice or GAI and RGA in Arabidopsis are repressors of plant development. DELLAs inhibit seed germination, seed growth, flowering and GA reverses these effects.[53] DELLA proteins are characterized by the presence of a DELLA motif (aspartate-glutamate-leucine-leucine-alanine or D-E-L-L-A in the single letter amino acid code).[54]
When GA binds to the GID1 receptor, it enhances the interaction between GID1 and DELLA proteins, forming a GA-GID1-DELLA complex. When in the GA-GID1-DELLA complex, it is thought that DELLA proteins undergo changes in structure that enable their binding to F-box proteins (SLY1 in Arabidopsis or GID2 in rice).[55][54][56] F-box proteins catalyse the addition of ubiquitin to their targets.[55] The addition of ubiquitin to DELLA proteins promotes their degradation via the 26S-proteosome.[54] The degradation of DELLA proteins releases cells from their repressive effects.
Transcription factors
The first targets of DELLA proteins identified were PHYTOCHROME INTERACTING FACTORs (PIFs). PIFs are transcription factors that negatively regulate light signalling and are strong promoters of elongation growth. In the presence of GA, DELLAs are degraded and this then allows PIFs to promote elongation.[57] It was later found that DELLAs repress a large number of other transcription factors, among which are positive regulators of auxin, brassinosteroid and ethylene signalling.[58][59] DELLAs can repress transcription factors either by stopping their binding to DNA or by promoting their degradation.[57]
Prefoldins and microtubule assembly
In addition to repressing transcription factors, DELLAs also bind to prefoldins (PFDs). PFDs are molecular chaperones, meaning they assist in the folding of other proteins. PFDs function in the cytosol but when DELLAs bind to PFDs, it restricts them to the nucleus. An important function of PFDs is to assist in the folding of β-tubulin. As such, in the absence of GA (when there is a high level of DELLA proteins), PDF function is reduced and there is a lower cellular pool of β-tubulin. When GA is present the DELLAs are degraded, PDFs can move to the cytosol and assist in the folding of β-tubulin. β-tubulin is a vital component of the cytoskeleton (in the form of microtubules). As such, GA allows for re-organisation of the cytoskeleton, and the elongation of cells.[60]
Microtubules are also required for the trafficking of membrane vesicles. Membrane vesicle trafficking is needed for the correct positioning of several hormone transporters. One of the most well characterized hormone transporters are PIN proteins, which are responsible for the movement of the hormone auxin between cells. In the absence of GA, DELLA proteins reduce the levels of microtubules and thereby inhibit membrane vesicle trafficking. This reduces the level of PIN proteins at the cell membrane, and the level of auxin in the cell. GA reverses this process and allows for PIN protein trafficking to the cell membrane to enhance the level of auxin in the cell.[61]
References
- Hedden P, Sponsel V (2015). "A Century of Gibberellin Research". Journal of Plant Growth Regulation. 34 (4): 740–60. doi:10.1007/s00344-015-9546-1. PMC 4622167. PMID 26523085.
- Spielmeyer W, Ellis MH, Chandler PM (June 2002). "Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene". Proceedings of the National Academy of Sciences of the United States of America. 99 (13): 9043–8. Bibcode:2002PNAS...99.9043S. doi:10.1073/pnas.132266399. PMC 124420. PMID 12077303.
- "Norman Borlaug: A Billion Lives Saved". www.agbioworld.org. Retrieved 2018-05-11.
- B B Stowe; Yamaki, T. (1957). "The History and Physiological Action of the Gibberellins". Annual Review of Plant Physiology. 8 (1): 181–216. doi:10.1146/annurev.pp.08.060157.001145.
- Mees, G.C.; Elson, G.W. (1978). "Chapter 7: The gibberellins". In Peacock, F.C. (ed.). Jealott's Hill: Fifty years of Agricultural Research 1928-1978. Imperial Chemical Industries Ltd. pp. 55–60. ISBN 0901747017.
- Gibberellin and Flame Seedless Grapes Archived 2006-12-06 at the Wayback Machine from a University of California, Davis website
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (April 2002). "Green revolution: a mutant gibberellin-synthesis gene in rice". Nature. 416 (6882): 701–2. Bibcode:2002Natur.416..701S. doi:10.1038/416701a. PMID 11961544. S2CID 4414560.
- Campbell N, Reec JB (2002). Biology (6th ed.). San Francisco: Benjamin Cummings. ISBN 9780805366242.
- Sponsel, Valerie M.; Hedden, Peter (2010), Davies, Peter J. (ed.), "Gibberellin Biosynthesis and Inactivation", Plant Hormones, Dordrecht: Springer Netherlands, pp. 63–94, doi:10.1007/978-1-4020-2686-7_4, ISBN 978-1-4020-2684-3, retrieved 2022-01-29
- Hedden, Peter (2020-11-23). "The Current Status of Research on Gibberellin Biosynthesis". Plant and Cell Physiology. 61 (11): 1832–1849. doi:10.1093/pcp/pcaa092. ISSN 1471-9053. PMC 7758035. PMID 32652020.
- "Gibberellins". AccessScience. doi:10.1036/1097-8542.289000.
- Yamaguchi S (2008). "Gibberellin metabolism and its regulation". Annual Review of Plant Biology. 59: 225–51. doi:10.1146/annurev.arplant.59.032607.092804. PMID 18173378.
- MacMillan J (December 2001). "Occurrence of Gibberellins in Vascular Plants, Fungi, and Bacteria". Journal of Plant Growth Regulation. 20 (4): 387–442. doi:10.1007/s003440010038. PMID 11986764. S2CID 44504525.
- Davies PJ. "Plant growth". AccessScience. doi:10.1036/1097-8542.523000.
- Hedden P, Thomas SG (May 2012). "Gibberellin biosynthesis and its regulation". The Biochemical Journal. 444 (1): 11–25. doi:10.1042/BJ20120245. PMID 22533671. S2CID 25627726.
- Koornneef M, van der Veen JH (November 1980). "Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh". Theoretical and Applied Genetics. 58 (6): 257–63. doi:10.1007/BF00265176. PMID 24301503. S2CID 22824299.
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (March 2006). "Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development". The Plant Journal. 45 (5): 804–18. doi:10.1111/j.1365-313X.2005.02642.x. PMID 16460513.
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T (December 1998). "Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds". The Plant Cell. 10 (12): 2115–26. doi:10.1105/tpc.10.12.2115. PMC 143973. PMID 9836749.
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (February 2004). "Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds". The Plant Cell. 16 (2): 367–78. doi:10.1105/tpc.018143. PMC 341910. PMID 14729916.
- Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P (March 1999). "Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes". The Plant Journal. 17 (5): 547–56. doi:10.1046/j.1365-313X.1999.00410.x. PMID 10205907.
- Huang S, Raman AS, Ream JE, Fujiwara H, Cerny RE, Brown SM (November 1998). "Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis". Plant Physiology. 118 (3): 773–81. doi:10.1104/pp.118.3.773. PMC 34787. PMID 9808721.
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M (July 2003). "Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants?". The Plant Journal. 35 (1): 104–15. doi:10.1046/j.1365-313X.2003.01780.x. PMID 12834406.
- Itoh H, Tanaka-Ueguchi M, Kawaide H, Chen X, Kamiya Y, Matsuoka M (October 1999). "The gene encoding tobacco gibberellin 3beta-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development". The Plant Journal. 20 (1): 15–24. doi:10.1046/j.1365-313X.1999.00568.x. PMID 10571861.
- Rojas MC, Hedden P, Gaskin P, Tudzynski B (May 2001). "The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis". Proceedings of the National Academy of Sciences of the United States of America. 98 (10): 5838–43. Bibcode:2001PNAS...98.5838R. doi:10.1073/pnas.091096298. PMC 33300. PMID 11320210.
- Kawaide H, Imai R, Sassa T, Kamiya Y (August 1997). "Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis". The Journal of Biological Chemistry. 272 (35): 21706–12. doi:10.1074/jbc.272.35.21706. PMID 9268298.
- Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, Sassa T (March 2000). "Cloning of a full-length cDNA encoding ent-kaurene synthase from Gibberella fujikuroi: functional analysis of a bifunctional diterpene cyclase". Bioscience, Biotechnology, and Biochemistry. 64 (3): 660–4. doi:10.1271/bbb.64.660. PMID 10803977.
- Tudzynski B, Kawaide H, Kamiya Y (September 1998). "Gibberellin biosynthesis in Gibberella fujikuroi: cloning and characterization of the copalyl diphosphate synthase gene". Current Genetics. 34 (3): 234–40. doi:10.1007/s002940050392. PMID 9745028. S2CID 3021994.
- Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B (December 2001). "Gibberellin Biosynthesis in Plants and Fungi: A Case of Convergent Evolution?". Journal of Plant Growth Regulation. 20 (4): 319–331. doi:10.1007/s003440010037. PMID 11986758. S2CID 25623658.
- Kawaide H (March 2006). "Biochemical and molecular analyses of gibberellin biosynthesis in fungi". Bioscience, Biotechnology, and Biochemistry. 70 (3): 583–90. doi:10.1271/bbb.70.583. PMID 16556972. S2CID 20952424.
- Lopes AL, Silva DN, Rodrigues C, Costa JL, Machado MP, Penha RO, Biasi LA, Ricardo C (2013). "Gibberellic acid fermented extract obtained by solid-state fermentation using citric pulp by Fusarium moniliforme: Influence on Lavandula angustifolia Mill. cultivated in vitro". Pak J Bot. 45: 2057–2064.
- Thomas SG, Phillips AL, Hedden P (April 1999). "Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation". Proceedings of the National Academy of Sciences of the United States of America. 96 (8): 4698–703. Bibcode:1999PNAS...96.4698T. doi:10.1073/pnas.96.8.4698. PMC 16395. PMID 10200325.
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM (January 2003). "Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants". The Plant Cell. 15 (1): 151–63. doi:10.1105/tpc.005975. PMC 143488. PMID 12509528.
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, Zhu X, Mander LN, Kamiya Y, Yamaguchi S, He Z (February 2006). "ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice". The Plant Cell. 18 (2): 442–56. doi:10.1105/tpc.105.038455. PMC 1356550. PMID 16399803.
- Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, Ma CJ, Noel JP, Mander L, Shulaev V, Kamiya Y, Rodermel S, Weiss D, Pichersky E (January 2007). "Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2". The Plant Cell. 19 (1): 32–45. doi:10.1105/tpc.106.044602. PMC 1820973. PMID 17220201.
- Hedden P, Phillips AL (December 2000). "Gibberellin metabolism: new insights revealed by the genes". Trends in Plant Science. 5 (12): 523–30. doi:10.1016/S1360-1385(00)01790-8. PMID 11120474.
- Olszewski N, Sun TP, Gubler F (2002). "Gibberellin signaling: biosynthesis, catabolism, and response pathways". The Plant Cell. 14 Suppl (Suppl): S61–80. doi:10.1105/tpc.010476. PMC 151248. PMID 12045270.
- Chiang HH, Hwang I, Goodman HM (February 1995). "Isolation of the Arabidopsis GA4 locus". The Plant Cell. 7 (2): 195–201. doi:10.1105/tpc.7.2.195. PMC 160775. PMID 7756830.
- Matsushita A, Furumoto T, Ishida S, Takahashi Y (March 2007). "AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase". Plant Physiology. 143 (3): 1152–62. doi:10.1104/pp.106.093542. PMC 1820926. PMID 17277098.
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (July 1995). "Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis". Plant Physiology. 108 (3): 1049–57. doi:10.1104/pp.108.3.1049. PMC 157456. PMID 7630935.
- Xu YL, Li L, Gage DA, Zeevaart JA (May 1999). "Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis". The Plant Cell. 11 (5): 927–36. doi:10.1105/tpc.11.5.927. PMC 144230. PMID 10330476.
- Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LH, Elliott RC (March 2000). "Evidence that auxin promotes gibberellin A1 biosynthesis in pea". The Plant Journal. 21 (6): 547–52. doi:10.1046/j.1365-313x.2000.00702.x. PMID 10758505.
- Wolbang CM, Ross JJ (November 2001). "Auxin promotes gibberellin biosynthesis in decapitated tobacco plants". Planta. 214 (1): 153–7. doi:10.1007/s004250100663. PMID 11762165. S2CID 31185063.
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ (February 2004). "Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems". Plant Physiology. 134 (2): 769–76. doi:10.1104/pp.103.030460. PMC 344552. PMID 14730077.
- Ngo P, Ozga JA, Reinecke DM (July 2002). "Specificity of auxin regulation of gibberellin 20-oxidase gene expression in pea pericarp". Plant Molecular Biology. 49 (5): 439–48. doi:10.1023/A:1015522404586. PMID 12090620. S2CID 22530544.
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP (April 2007). "The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes". Proceedings of the National Academy of Sciences of the United States of America. 104 (15): 6484–9. Bibcode:2007PNAS..104.6484A. doi:10.1073/pnas.0610717104. PMC 1851083. PMID 17389366.
- Vettakkorumakankav NN, Falk D, Saxena P, Fletcher RA (1999). "A Crucial Role for Gibberellins in Stress Protection of Plants". Plant and Cell Physiology. 40 (5): 542–548. doi:10.1093/oxfordjournals.pcp.a029575.
- Batge SL, Ross JJ, Reid JB (1999). "Abscisic acid levels in seeds of the gibberellin-deficient mutant lh-2 of pea (Pisum sativum)". Physiologia Plantarum. 195 (3): 485–490. doi:10.1034/j.1399-3054.1999.105313.x.
- White CN, Proebsting WM, Hedden P, Rivin CJ (April 2000). "Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways". Plant Physiology. 122 (4): 1081–8. doi:10.1104/pp.122.4.1081. PMC 58942. PMID 10759503.
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (September 2004). "The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid". Developmental Cell. 7 (3): 373–85. doi:10.1016/j.devcel.2004.06.017. PMID 15363412.
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, Matsuoka M (July 2007). "Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin". The Plant Cell. 19 (7): 2140–55. doi:10.1105/tpc.106.043729. PMC 1955699. PMID 17644730.
- Murase K, Hirano Y, Sun TP, Hakoshima T (November 2008). "Gibberellin-induced DELLA recognition by the gibberellin receptor GID1". Nature. 456 (7221): 459–63. Bibcode:2008Natur.456..459M. doi:10.1038/nature07519. PMID 19037309. S2CID 16280595.
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M (November 2008). "Structural basis for gibberellin recognition by its receptor GID1". Nature. 456 (7221): 520–3. Bibcode:2008Natur.456..520S. doi:10.1038/nature07546. PMID 19037316. S2CID 205215510.
- Achard P, Genschik P (2009). "Releasing the brakes of plant growth: how GAs shutdown DELLA proteins". Journal of Experimental Botany. 60 (4): 1085–92. doi:10.1093/jxb/ern301. PMID 19043067.
- Davière JM, Achard P (March 2013). "Gibberellin signaling in plants". Development. 140 (6): 1147–51. doi:10.1242/dev.087650. PMID 23444347.
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P (December 2006). "F-box proteins everywhere". Current Opinion in Plant Biology. 9 (6): 631–8. doi:10.1016/j.pbi.2006.09.003. PMID 17005440.
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM (May 2003). "The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase". The Plant Cell. 15 (5): 1120–30. doi:10.1105/tpc.010827. PMC 153720. PMID 12724538.
- Zheng Y, Gao Z, Zhu Z (October 2016). "DELLA-PIF Modules: Old Dogs Learn New Tricks". Trends in Plant Science. 21 (10): 813–815. doi:10.1016/j.tplants.2016.08.006. PMID 27569991.
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (May 2014). "Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl". eLife. 3. doi:10.7554/eLife.03031. PMC 4075450. PMID 24867218.
- Marín-de la Rosa N, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñate-Sánchez L, Holdsworth MJ, Bhalerao R, Alabadí D, Blázquez MA (October 2014). "Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners". Plant Physiology. 166 (2): 1022–32. doi:10.1104/pp.114.244723. PMC 4213073. PMID 25118255.
- Locascio A, Blázquez MA, Alabadí D (May 2013). "Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction". Current Biology. 23 (9): 804–9. doi:10.1016/j.cub.2013.03.053. PMID 23583555.
- Salanenka Y, Verstraeten I, Löfke C, Tabata K, Naramoto S, Glanc M, Friml J (April 2018). "Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane". Proceedings of the National Academy of Sciences of the United States of America. 115 (14): 3716–3721. doi:10.1073/pnas.1721760115. PMC 5889667. PMID 29463731.
External links
- Gibberellin in the Pesticide Properties DataBase (PPDB)