Induced cell cycle arrest
Induced cell cycle arrest is the use of a chemical or genetic manipulation to artificially halt progression through the cell cycle. Cellular processes like genome duplication and cell division stop.[1] It can be temporary or permanent.[1] It is an artificial activation of naturally occurring cell cycle checkpoints, induced by exogenous stimuli controlled by an experimenter.
Model organisms
In an academic research context, cell cycle arrest is typically performed in model organisms and cell extracts, such as Saccharomyces cervisiae (yeast) or Xenopus oocytes (frog eggs).[2][3] Frog egg cell extracts have been used extensively in cell cycle research because they are relatively large, reaching a diameter of 1mm, and so contain large amounts of protein, making protein levels more easily measurable.[4]
Purposes
There are a variety of reasons a researcher may want to temporarily or permanently prevent progress through the cell cycle.
Cell cycle synchronization
In some experiments, a researcher may want to control and synchronize the time when a group of cells progress to the next phase of the cell cycle.[5] The cells can be induced to arrest as they arrive (at different time points) at a certain phase, so that when the arrest is lifted (for instance, rescuing cell cycle progression by introducing another chemical) all the cells resume cell cycle progression at the same time. In addition to this method acting as a scientific control for when the cells resume the cell cycle, this can be used to investigate necessity and sufficiency.
Another reason synchrony is important is the control for amount of DNA content, which varies at different parts of the cell cycle based on whether DNA replication has occurred since the last round of completed mitosis and cytokinesis.[6]
Furthermore, synchronization of large numbers of cells into the same phase allows for the collection of large enough groups of cells in the same cycle for the use in other assays, such as western blot and RNA sequencing.[7]
DNA damage repair
Researchers may be investigating mechanisms of DNA damage repair. Given that some of the mechanisms below of inducing cell cycle arrest involve damaging the DNA, this allows investigation into how the cell responds to damage of its genetic material.[8]
Identification of in vivo protein function
Genetic engineering of cells with specific gene knockouts can also result in cells that arrest at different phases of the cell cycle. Examples include:
- G1: Saccharomyces cerevisiae yeast expressing dominant mutant alleles of CDC28 arrest in G1, which indicates that CDC28 is necessary for passage beyond the G1 phase.[9]
- S: Schizosaccharomyces pombe (fission yeast) expressing a temperature-sensitive mutant form of DNA polymerase delta (pol delta ts03) arrest in S phase.[10]
- G2: Fission yeast expressing some mutant forms of CDC2 unable to arrest in G2 in response to DNA damage, indicating the gene product is involved in G2 arrest.[11]
- M: A mutant screen of budding yeasts with mitotic arrest identified CDC16, CDC23, and CDC27 as key genes that, when mutated, cause arrest in mitosis.[12]
G1 phase arrest
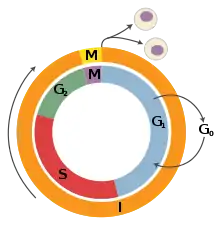
G1 phase is the first of the four phases of the cell cycle, and is part of interphase. While in G1 the cell synthesizes messenger RNA (mRNA) and proteins in preparation for subsequent steps of interphase leading to mitosis. In human somatic cells, the cell cycle lasts about 18 hours, and the G1 phase makes up about 1/3 of that time.[13] On the other hand, in frog, sea urchin, and fruit fly embryos, the G1 phase is extremely brief and instead is a slight gap between cytokinesis and S phase.[13]
Alpha Factor
α-factor is a pheromone secreted by Saccharomyces cervisiae that arrests the yeast cells in G1 phase. It does so by inhibiting the enzyme adenylate cyclase.[2] The enzyme catalyzes the conversion of adenosine triphosphate (ATP) to 3',5'-cyclic AMP (cAMP) and pyrophosphate.[14]
Contact inhibition
Contact inhibition is a method of arresting cells when neighboring cells come into contact with each other. It results in a single layer of arrested cells of arrested cells, and is a process that is notably missing in cancer cells. The suspected mechanism is dependent on p27Kip1, a cyclin-dependent kinase inhibitor.[15] p27Kip1 protein levels are elevated in arresting cells. This natural process can be mimicked in a lab through the overexpression of p27Kip1, which results in induced cell cycle arrest in G1 phase.[16]
Mimosine
Mimosine is a plant amino acid that has been shown to reversibly inhibit progression beyond G1 phase in some human cells, including lymphoblastoid cells.[5] Its proposed mechanism of action is an iron/zinc chelator that depletes iron within the cell. This induces double-strand breaks in the DNA, inhibiting DNA replication. This may involve blocking the action of an iron-dependent ribonucleotide reductase. It may also inhibit transcription of serine hydroxymethyltransferase, which has zinc dependence.[17]
Serum deprivation
In cell culture, serum is the growth medium in which the cells are grown and contains vital nutrients. The use of serum deprivation - partially or completely removing the serum and its nutrients - has been shown to arrest and synchronize cell cycle progression in G0 phase, for example in neonatal mammalian astrocytes[18] and human foreskin fibroblasts.[19]
Amino acid starvation is a similar approach. When grown in a media without some essential amino acids, such as methionine, some cells arrest in early G1 phase.[5]
S phase arrest
S phase follows G1 phase via the G1/S transition and precedes G2 phase in interphase and is the part of the cell cycle in which DNA is replicated. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved. Pre-replication complexes assembled before S phase are converted into active replication forks.[20] Driving this conversion is Cdc7 and S-phase cyclin-dependent kinases, which are both upregulated after the G1/S transition.[20]
Aphidicolin
Aphidicolin is an antibiotic isolated from the fungus Cephalosporum aphidicola. It is a reversible inhibitor of eukaryotic nuclear DNA replication that blocks progression past the S phase. Its mechanism is the inhibition of DNA polymerase A and D. A structural study found that this is thought to occur through binding the alpha active site of the polymerase and "rotating the template guanine," which prevents deoxycytidine triphosphate (dCTP) from binding.[21] This S phase block induces apoptosis in HeLa cells.[5]
Hydroxyurea
Hydroxyurea (HU) is a small molecule drug that inhibits the enzyme ribonucleotide reductase (RNR), preventing the catalysis of converting deoxyribonucleotides (DNTs) to ribonucleotides. It is hypothesized that there is tyrosyl free radical within RNR that is disabled by HU.[6][22] The free radicals are necessary for the reduction of the DNTs and are scavenged by HU instead.[23] HU has been shown to arrest cells in both S phase (healthy cells) and immediately before cytokinesis (mutant cells).[22]
2,3-DCPE
2[[3-(2,3-dichlorophenoxy)propyl] amino]ethanol (2,3-DCPE) is a small-molecule that induces S phase arrest.[24] This was demonstrated in cancer cell lines and downregulates expression of B-cell lymphoma-extra large (Bcl-XL), an anti-apoptotic protein that prevents the release of mitochondrial contents like cytochrome c.
G2 phase arrest
G2 phase is the final part of interphase and directly precedes mitosis. It will only be entered in regular cells if the DNA replication in S phase is completed successfully. It is a period of rapid cell growth and protein synthesis during which the cell prepares itself for mitosis.
Destruction of cyclin mRNA
Cyclins are proteins that control progression through the cell cycle by activating cyclin-dependent kinases. Destruction of a cell's endogenous cyclin messenger RNA can arrest frog egg extracts in interphase and prevent them from entering mitosis.[3] Introduction of exogenous cyclin mRNA is also sufficient to rescue cell cycle progression.[3] One method of this destruction is through the use of antisense oligonucleotides, pieces of RNA that bind to the cyclin mRNA and prevent the mRNA from being translated into cyclin protein.[25] This can actually be used to destroy phase-specific cyclins beyond just G2 - for instance, destruction of cyclin D1 mRNA by antisense oligonucleotides prevents progression from G1 phase to S phase.[26]
Mitotic arrest
Mitosis is the final part of the cell cycle and follows interphase. It is composed of four phases - prophase, metaphase, anaphase, and telophase - and involves the condensation of the chromosomes in the nucleus, the dissolution of the nuclear envelope, and the separation of sister chromatids by spindle fibers. As mitosis concludes, the spindle fibers disappear and the nuclear membrane reforms around each of the two sets of chromosomes. After successful mitosis, the cell physically splits into two identical daughter cells in a process called cytokinesis, and this concludes a full round of the cell cycle. Each of these new cells could then potentially re-enter G1 phase and begin the cell cycle again.[27]
Nocodazole
Nocodazole is a chemical agent that interferes with the polymerization of microtubules.[28] Cells treated with nocodazole arrest with a G2 or M phase DNA content, which can be verified with flow cytometry. From microscopy it has been determined they do enter mitosis but they cannot form the spindles necessary for metaphase because the microtubules cannot polymerize.[29] Research into the mechanism has hinted at it potentially preventing tubulin from forming its alpha/beta heterodimer.[30]
Taxol
Taxol works in the opposite way of nocodazole, instead stabilizing the microtubule polymer and preventing it from disassembly. It also causes M phase arrest, as the spindle that is supposed to pull apart sister chromatids is unable to disassemble.[31][32] It acts through a specific binding site on the microtubule polymer, and as such does not require GTP or other cofactors to induce tubulin polymerization.[33]
Temperature
Temperature has been shown to regulate HeLa cell cycle progression. Mitosis was found to be the most temperature-sensitive part of the cell cycle.[34] Pre-cytokinesis mitotic arrest was visible through accumulation of cells in mitosis in below-normal temperatures between 24 and 31 °C (75.2-87.8 °F).[34]
Verification
There are several methods that can be used to verify that cells have been arrested in the proper phase.
Flow cytometry
Flow cytometry is a technique of measuring physical and chemical characteristics of a population of cells using lasers and fluorophore dyes covalently linked to protein markers.[35] The stronger the signal, the more of a particular protein is present. Staining with DNA dyes propidium iodide or 4',6'-diamidino-2-phenylindole (DAPI) allows delineation or sorting of cells between G1, S, or G2/M phases.[36]
Immunoblotting
Immunoblotting is the detection of specific proteins in a tissue sample or extract. Primary antibodies recognize and bind the protein in question, and secondary antibodies are added that recognize the primary antibodies. The secondary antibody is then visualized through staining or immunofluorescence, allowing indirect detection of the original target protein.
Immunoblotting can be performed to detect the presence of cyclins, proteins that regulate the cell cycle.[37] Different classes of cyclins are up- and down-regulated at different parts of the cell cycle. Measurement of the cyclins from an extract of an arrested cell can determine what phase the cell is in. For example, a peak of cyclin E protein would indicate the G1/S transition, a cyclin A peak would indicate late G2 phase, and a cyclin B peak would indicate mitosis.[38]
Fluorescence ubiquitination-based cell cycle indication (FUCCI)
FUCCI is a system that takes advantage of cell cycle phase-specific expression of proteins and their degradation by the ubiquitin-proteasome pathway. Two fluorescent probes - Cdt1 and Geminin conjugated to fluorescent proteins - allow for real-time visualization of the cell cycle phase a cell is in.[39]
References
- Li Y, Fan J, Ju D (Jan 1, 2019). "15 - Neurotoxicity concern about the brain targeting delivery systems". In Gao H, Gao X (eds.). Brain Targeted Drug Delivery System. Academic Press. pp. 377–408. doi:10.1016/B978-0-12-814001-7.00015-9. ISBN 978-0-12-814001-7. S2CID 105749019.
- Liao H, Thorner J (April 1980). "Yeast mating pheromone alpha factor inhibits adenylate cyclase". Proceedings of the National Academy of Sciences of the United States of America. 77 (4): 1898–902. Bibcode:1980PNAS...77.1898L. doi:10.1073/pnas.77.4.1898. PMC 348616. PMID 6246513.
- Murray AW, Kirschner MW (May 1989). "Cyclin synthesis drives the early embryonic cell cycle". Nature. 339 (6222): 275–80. Bibcode:1989Natur.339..275M. doi:10.1038/339275a0. PMID 2566917. S2CID 4352582.
- Lodish H, Berk A, Zipursky SL, et al. (2000). "Section 13.2: Biochemical Studies with Oocytes, Eggs, and Early Embryos". Molecular Cell Biology (4th ed.). New York: W. H. Freeman. ISBN 0-7167-3136-3.
- Krek W, DeCaprio JA (1995). "[7] Cell synchronization". Cell synchronization. Methods in Enzymology. Vol. 254. Elsevier. pp. 114–24. doi:10.1016/0076-6879(95)54009-1. ISBN 978-0-12-182155-5. PMID 8531680.
- Koç A, Wheeler LJ, Mathews CK, Merrill GF (January 2004). "Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools". The Journal of Biological Chemistry. 279 (1): 223–30. doi:10.1074/jbc.M303952200. PMID 14573610.
- Purcell M, Kruger A, Tainsky MA (2014-12-15). "Gene expression profiling of replicative and induced senescence". Cell Cycle. 13 (24): 3927–37. doi:10.4161/15384101.2014.973327. PMC 4615143. PMID 25483067.
- Singh A, Xu YJ (November 2016). "The Cell Killing Mechanisms of Hydroxyurea". Genes. 7 (11): 99. doi:10.3390/genes7110099. PMC 5126785. PMID 27869662.
- Mendenhall MD, Richardson HE, Reed SI (June 1988). "Dominant negative protein kinase mutations that confer a G1 arrest phenotype". Proceedings of the National Academy of Sciences of the United States of America. 85 (12): 4426–30. Bibcode:1988PNAS...85.4426M. doi:10.1073/pnas.85.12.4426. PMC 280442. PMID 3288995.
- Uchiyama M, Galli I, Griffiths DJ, Wang TS (June 1997). "A novel mutant allele of Schizosaccharomyces pombe rad26 defective in monitoring S-phase progression to prevent premature mitosis". Molecular and Cellular Biology. 17 (6): 3103–15. doi:10.1128/MCB.17.6.3103. PMC 232163. PMID 9154809.
- al-Khodairy F, Carr AM (April 1992). "DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe". The EMBO Journal. 11 (4): 1343–50. doi:10.1002/j.1460-2075.1992.tb05179.x. PMC 556583. PMID 1563350.
- Hwang LH, Murray AW (October 1997). "A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis". Molecular Biology of the Cell. 8 (10): 1877–87. doi:10.1091/mbc.8.10.1877. PMC 25633. PMID 9348530.
- Morgan DO (2007). The cell cycle : principles of control. New Science Press. ISBN 978-0-19-920610-0. OCLC 70173205.
- Zhang G, Liu Y, Ruoho AE, Hurley JH (March 1997). "Structure of the adenylyl cyclase catalytic core". Nature. 386 (6622): 247–53. Bibcode:1997Natur.386..247Z. doi:10.1038/386247a0. PMID 9069282. S2CID 4329051.
- Seluanov A, Hine C, Azpurua J, Feigenson M, Bozzella M, Mao Z, et al. (November 2009). "Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat". Proceedings of the National Academy of Sciences of the United States of America. 106 (46): 19352–7. Bibcode:2009PNAS..10619352S. doi:10.1073/pnas.0905252106. PMC 2780760. PMID 19858485.
- Li J, Yang XK, Yu XX, Ge ML, Wang WL, Zhang J, Hou YD (August 2000). "Overexpression of p27(KIP1) induced cell cycle arrest in G(1) phase and subsequent apoptosis in HCC-9204 cell line". World Journal of Gastroenterology. 6 (4): 513–521. doi:10.3748/wjg.v6.i4.513 (inactive 1 August 2023). PMC 4723549. PMID 11819639.
{{cite journal}}: CS1 maint: DOI inactive as of August 2023 (link) - "Mimosine". www.drugbank.ca. Retrieved 2019-12-13.
- Langan TJ, Rodgers KR, Chou RC (2016-11-05). "Synchronization of Mammalian Cell Cultures by Serum Deprivation". Cell Cycle Synchronization. Methods in Molecular Biology. Vol. 1524. Springer New York. pp. 97–105. doi:10.1007/978-1-4939-6603-5_6. ISBN 978-1-4939-6602-8. PMID 27815898.
- Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF (June 2014). "Cyclin D activates the Rb tumor suppressor by mono-phosphorylation". eLife. 3: e02872. doi:10.7554/eLife.02872. PMC 4076869. PMID 24876129.
- Takeda DY, Dutta A (April 2005). "DNA replication and progression through S phase". Oncogene. 24 (17): 2827–43. doi:10.1038/sj.onc.1208616. PMID 15838518.
- Baranovskiy AG, Babayeva ND, Suwa Y, Gu J, Pavlov YI, Tahirov TH (December 2014). "Structural basis for inhibition of DNA replication by aphidicolin". Nucleic Acids Research. 42 (22): 14013–21. doi:10.1093/nar/gku1209. PMC 4267640. PMID 25429975.
- Xu YJ, Singh A, Alter GM (November 2016). "Hydroxyurea Induces Cytokinesis Arrest in Cells Expressing a Mutated Sterol-14α-Demethylase in the Ergosterol Biosynthesis Pathway". Genetics. 204 (3): 959–973. doi:10.1534/genetics.116.191536. PMC 5105871. PMID 27585850.
- Platt OS (March 2008). "Hydroxyurea for the treatment of sickle cell anemia". The New England Journal of Medicine. 358 (13): 1362–9. doi:10.1056/NEJMct0708272. PMID 18367739.
- Zhu H, Zhang L, Wu S, Teraishi F, Davis JJ, Jacob D, Fang B (June 2004). "Induction of S-phase arrest and p21 overexpression by a small molecule 23-(2,3-dichlorophenoxy)propyl] amino]ethanol in correlation with activation of ERK". Oncogene. 23 (29): 4984–92. doi:10.1038/sj.onc.1207645. PMID 15122344.
- "NCI Dictionary of Cancer Terms". National Cancer Institute. 2011-02-02. Retrieved 2019-12-13.
- Saikawa Y, Kubota T, Otani Y, Kitajima M, Modlin IM (October 2001). "Cyclin D1 antisense oligonucleotide inhibits cell growth stimulated by epidermal growth factor and induces apoptosis of gastric cancer cells". Japanese Journal of Cancer Research. 92 (10): 1102–9. doi:10.1111/j.1349-7006.2001.tb01065.x. PMC 5926617. PMID 11676861.
- "Mitosis - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2019-12-12.
- Kuhn M (March 1998). "The microtubule depolymerizing drugs nocodazole and colchicine inhibit the uptake of Listeria monocytogenes by P388D1 macrophages". FEMS Microbiology Letters. 160 (1): 87–90. doi:10.1111/j.1574-6968.1998.tb12895.x. PMID 9495017.
- Kanthou C, Tozer GM (June 2009). "Microtubule depolymerizing vascular disrupting agents: novel therapeutic agents for oncology and other pathologies". International Journal of Experimental Pathology. 90 (3): 284–94. doi:10.1111/j.1365-2613.2009.00651.x. PMC 2697551. PMID 19563611.
- Xu K, Schwarz PM, Ludueña RF (Feb 2002). "Interaction of nocodazole with tubulin isotypes". Drug Development Research. 55 (2): 91–96. doi:10.1002/ddr.10023. ISSN 0272-4391. S2CID 86235729.
- Choi YH, Yoo YH (December 2012). "Taxol-induced growth arrest and apoptosis is associated with the upregulation of the Cdk inhibitor, p21WAF1/CIP1, in human breast cancer cells". Oncology Reports. 28 (6): 2163–9. doi:10.3892/or.2012.2060. PMID 23023313.
- Ikui AE, Yang CP, Matsumoto T, Horwitz SB (October 2005). "Low concentrations of taxol cause mitotic delay followed by premature dissociation of p55CDC from Mad2 and BubR1 and abrogation of the spindle checkpoint, leading to aneuploidy". Cell Cycle. 4 (10): 1385–8. doi:10.4161/cc.4.10.2061. PMID 16138009.
- Horwitz SB (1994). "Taxol (paclitaxel): mechanisms of action". Annals of Oncology. 5 (Suppl 6): S3-6. PMID 7865431.
- Rao PN, Engelberg J (May 1965). "He La Cells: Effects of Temperature on the Life Cycle". Science. 148 (3673): 1092–4. Bibcode:1965Sci...148.1092R. doi:10.1126/science.148.3673.1092. PMID 14289609. S2CID 27085343.
- Picot J, Guerin CL, Le Van Kim C, Boulanger CM (March 2012). "Flow cytometry: retrospective, fundamentals and recent instrumentation". Cytotechnology. 64 (2): 109–30. doi:10.1007/s10616-011-9415-0. PMC 3279584. PMID 22271369.
- Pozarowski P, Darzynkiewicz Z (2004-07-01). "Analysis of cell cycle by flow cytometry". Checkpoint Controls and Cancer. Methods in Molecular Biology. Vol. 281. Humana Press. pp. 301–11. doi:10.1385/1-59259-811-0:301. ISBN 978-1-59259-811-3. PMID 15220539.
- Jenkins CW, Xiong Y (1996). "Immunoprecipitation and Immunoblotting in Cell Cycle Studies". Cell Cycle — Materials and Methods. Springer Berlin Heidelberg. pp. 250–263. doi:10.1007/978-3-642-57783-3_22. ISBN 978-3-540-58066-9.
- "Cyclin - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2019-12-12.
- "Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI)". MBL International. Retrieved 2019-12-12.