Marine microbiome
All animals on Earth form associations with microorganisms, including protists, bacteria, archaea, fungi, and viruses. In the ocean, animal–microbial relationships were historically explored in single host–symbiont systems. However, new explorations into the diversity of marine microorganisms associating with diverse marine animal hosts is moving the field into studies that address interactions between the animal host and a more multi-member microbiome. The potential for microbiomes to influence the health, physiology, behavior, and ecology of marine animals could alter current understandings of how marine animals adapt to change, and especially the growing climate-related and anthropogenic-induced changes already impacting the ocean environment.[1]
In the oceans, it is challenging to find eukaryotic organisms that do not live in close relationship with a microbial partner. Host-associated microbiomes also influence biogeochemical cycling within ecosystems with cascading effects on biodiversity and ecosystem processes. The microbiomes of diverse marine animals are currently under study, from simplistic organisms including sponges and ctenophores to more complex organisms such as sea squirts and sharks.
Background
| Part of a series on |
| Microbiomes |
|---|
 |
|


Within the vast biological diversity that inhabits the world's oceans, it would be challenging to find a eukaryotic organism that does not live in close relationship with a microbial partner.[2] Such symbioses, i.e., persistent interactions between host and microbe in which none of the partners gets harmed and at least one of them benefits, are ubiquitous from shallow reefs to deep-sea hydrothermal vents. Studies on corals,[3] sponges,[4] and mollusks [5][6][7] have revealed some of the profoundly important symbiotic roles microbes play in the lives of their hosts. These studies, however, have tended to focus on a small number of specific microbial taxa. In contrast, most hosts retain groups of many hundreds of different microbes (i.e., a microbiome,[8][9] which themselves can vary throughout the ontogeny of the host and as a result of environmental perturbations.[10][11][12] Rather than host-associated microbes functioning independently, complex multi-assemblage microbiomes have major impact on the fitness and function of their hosts. Studying these complex interactions and biological outcomes is difficult, but to understand the origin and evolution of organisms and populations and the structure and function of communities and ecosystems, the understanding of symbioses in host–microbiome systems needs advancing.[13][14][2]
There are many outstanding questions in ecology and evolution that could be addressed by expanding the phylogenetic and ecological breadth of host-associated microbiome studies, including all possible interactions throughout the microbiome. There is strong empirical evidence and new consensus that biodiversity (i.e., the richness of species and their interactions) pervasively influences the functioning of Earth's ecosystems, including ecosystem productivity.[15][16] However, this research has focused almost exclusively on macroorganisms. Because microbial symbionts are integral parts of most living organisms,[11][17] the understanding of how microbial symbionts contribute to host performance and adaptability needs broadening.[2]
Foundations of productive ecosystems
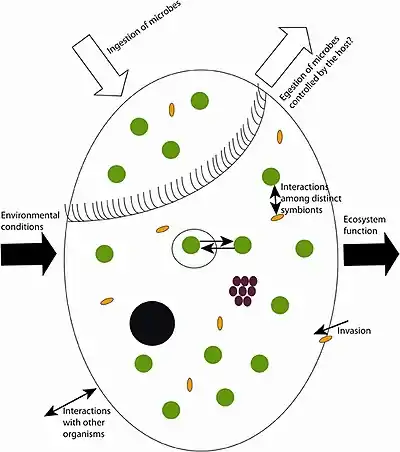
Black circle: macronucleus, white big circle: food vacuoles, green circles: phototrophs, brown circles: chemoautotrophs, yellow ovals: heterotrophic prokaryotes
Ecosystem engineers, such as many types of corals, deep-sea mussels, and hydrothermal vent tubeworms, contribute to primary productivity and create the structural habitats and nutrient resources that are the foundation of their respective ecosystems.[18] All of these taxa engage in mutualistic nutritional symbioses with microbes. There are many examples of marine nutritional mutualisms in which microbes enable hosts to utilize resources or substrates otherwise unavailable to the host alone. Such symbioses have been described in detail in reduced and anoxic sediments (e.g., lucinid clams, stilbonematid nematodes, and gutless oligochaetes) and hydrothermal vents (e.g., the giant tube worm or deep-sea mussels).[19] Moreover, many foundational species of marine macroalgae are vitamin auxotrophs (for example, half of more than 300 surveyed species were unable to synthesize cobalamin), and their productivity depends on provisioning from their epiphytic bacteria.[20] Reefs often consist of stony corals, one of the most well-known examples of a mutualistic symbiosis, in which the dinoflagellate alga Symbiodiniaceae supplies the coral with glucose, glycerol, and amino acids, while the coral provides the algae with a protected environment and limiting compounds (e.g., nitrogen species) needed for photosynthesis. However, this is a classic example of a mutualistic symbiosis that is sensitive to environmental disturbances, which can disrupt the fragile interactions between host and microbe. When reefs become warm and eutrophic, mutualistic Symbiodiniaceae may induce cellular damage to the host and/or sequester more resources for their own growth, thereby injuring and parasitizing their hosts.[21][22] Reef fishes, which seek homes on coral reefs, are important in fostering coral recovery in the wake of disturbance. Epulopiscium bacteria in the guts of surgeonfishes produce enzymes that allow their hosts to digest complex polysaccharides, enabling the host fish to feed on tough, leathery red and brown macroalgae.[23] This trophic innovation has facilitated niche diversification among coral reef herbivores. Surgeonfishes are critical to the functioning of Indo-Pacific coral reefs, as they are among the only fishes capable of consuming large macroalgae that bloom in the wake of ecosystem disturbance and suppress coral recovery.[24][2]

Along with more standard examples of nutritional symbioses in animals, recent advances in genome sequencing technology have led to the discovery of many endosymbiotic associations in marine protists (a protist is a general term to refer to a non-monophyletic collection of unicellular eukaryotes that are not fungi or in the Plantae group) These illustrate the incorporation of various new biochemical functions, such as photosynthesis, nitrogen fixation and recycling, and methanogenesis, into protist hosts by endosymbionts.[25] Endosymbiosis in protists is widespread and represents an important source of innovation. Previously unrecognized metabolic innovations of marine microbial symbioses that are ecologically important are discovered regularly.[26] For example, Candidatus Kentron (a clade of Gammaproteobacteria found in association with ciliates) nourish their ciliate hosts in the genus Kentrophoros and recycle acetate and propionate, which are low-value cellular waste products from their hosts, into biomass.[27] Another example is the anaerobic marine ciliate Strombidium purpureum.[28] The ciliate lives under anaerobic conditions and harbors endosymbiotic purple nonsulfur bacteria that contain both bacteriochlorophyll a and spirilloxanthin. The endosymbionts are photosynthetically active; hence, this symbiosis represents an evolutionary transition of an aerobic organism to an anaerobic one while incorporating organelles.[2]
Reproduction and host development
| Part of a series of overviews on |
| Marine life |
|---|
 |
Extending beyond nutritional symbioses, microbial symbionts can alter the reproduction, development, and growth of their hosts. Specific bacterial strains in marine biofilms often directly control the recruitment of planktonic larvae and propagules, either by inhibiting settlement or by serving as a settlement cue.[29][30] For example, the settlement of zoospores from the green alga Ulva intestinalis onto the biofilms of specific bacteria is mediated by their attraction to the quorum-sensing molecule, acyl-homoserine lactone, secreted by the bacteria.[31] Classic examples of marine host–microbe developmental dependence include the observation that algal cultures grown in isolation exhibited abnormal morphologies [32] and the subsequent discovery of morphogenesis-inducing compounds, such as thallusin, secreted by epiphytic bacterial symbionts.[33] Bacteria are also known to influence the growth of marine plants, macroalgae, and phytoplankton by secreting phytohormones such as indole acetic acid and cytokinin-type hormones.[34][35][36] In the marine choanoflagellate Salpingoeca rosetta, both multicellularity and reproduction are triggered by specific bacterial cues, offering a view into the origins of bacterial control over animal development (reviewed by Woznica and King.[37] The benefit to the bacteria, in return, is that they receive physical space to colonize at particular points in the water column typically accessible only to planktonic microbes. Perhaps the best-studied example of intimate host–microbe interactions controlling animal development is the Hawaiian bobtail squid Euprymna scolopes.[38] It lives in a mutualistic symbiosis with the bioluminescent bacteria Aliivibrio fischeri. The bacteria are fed a solution of sugars and amino acids by the host and, in return, provide bioluminescence for countershading and predator avoidance.[7] This mutualism with microbes provides a selective advantage for the squid in predator–prey interactions. Another invertebrate example can be found in tubeworms, in which Hydroides elegans metamorphosis is mediated by a bacterial inducer and mitogen-activated protein kinase (MAPK) signaling in biofilms.[39][2]
Biofouling and microbial community assembly
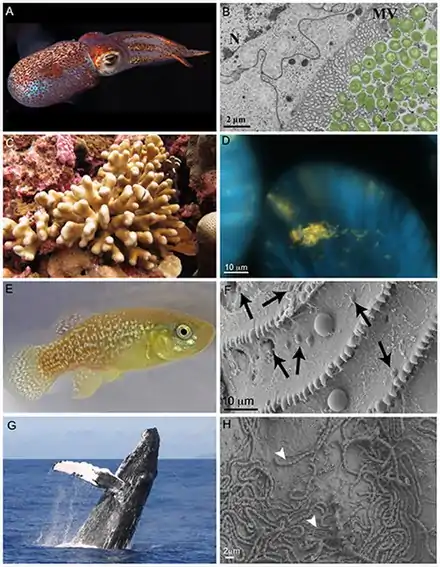
(C) the reef-building coral Stylophora pistillata and (D) a microscopy image of Endozoicomonas cells (probed yellow using in situ hybridization) within the tentacles of a S. pistillata host.
(E) the Atlantic killifish and (F) a SEM image of the surface and scales of the fish, with arrows pointing to bacterial-sized cells and larger cells (which are not noted) are presumably phytoplankton.
(G) a humpback whale breaching and (H) a SEM image of a humpback's skin surface associated bacteria, with arrows indicating two different cell morphologies.[1]
Some host-associated microbes produce compounds that prevent biofouling and regulate microbiome assembly and maintenance in many marine organisms, including sponges, macroalgae, and corals.[40][41] For example, tropical corals harbor diverse bacteria in their surface mucus layer that produce quorum-sensing inhibitors and other antibacterial compounds as a defense against colonization and infection by potential microbial pathogens.[3] Epiphytic bacteria of marine macroalgae excrete a diverse chemical arsenal capable of selectively shaping further bacterial colonization and deterring the settlement of biofouling marine invertebrates such as bryozoans.[34][42] As in corals, these diverse, microbially secreted compounds include not only bactericidal and bacteriostatic antibiotics but also compounds like halogenated furanones, cyclic dipeptides, and acyl-homoserine lactone mimics that disrupt bacterial quorum sensing and inhibit biofilm formation.[43] The bacteria likely are able to utilize the carbon-rich exudates from their hosts.[44][45] For example, in the case of giant kelp, the alga emits approximately 20% of primary production as dissolved organic carbon.[45] Whereas these prior examples illustrate how the microbiomes can protect hosts from surface colonization, a similar phenomenon has also been observed internally in the shipworm Bankia setacea, in which symbionts produce a boronated tartrolon antibiotic thought to keep the wood-digesting cecum clear of bacterial foulants.[46] By producing antimicrobial compounds, these microbes are able to defend their niche space to prevent other organisms from crowding them out.[2]
Biogeochemical cycling
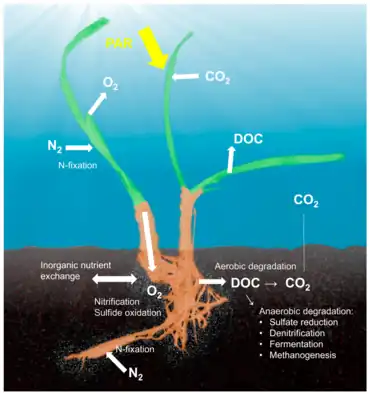
Host-associated microbiomes also influence biogeochemical cycling within ecosystems with cascading effects on biodiversity and ecosystem processes. For example, microbial symbionts comprise up to 40% of the biomass of their sponge hosts.[47] Through a process termed the "sponge-loop," they convert dissolved organic carbon released by reef organisms into particulate organic carbon that can be consumed by heterotrophic organisms.[4] Along with the coral–Symbiodiniaceae mutualism, this sponge-bacterial symbiosis helps explain Darwin's paradox, i.e., how highly productive coral reef ecosystems exist within otherwise oligotrophic tropical seas. Some sponge symbionts play a significant role in the marine phosphorus cycle by sequestering nutrients in the form of polyphosphate granules in the tissue of their host [48] and nitrogen cycling, e.g., through nitrification, denitrification, and ammonia oxidation.[4][41]]. Many macroalgal-associated bacteria are specifically adapted to degrade complex algal polysaccharides (e.g., fucoidan, porphyran, and laminarin [49][50]) and modify both the quality and quantity of organic carbon supplied to the ecosystem.[44][51] The sulfur-oxidizing gill endosymbionts of lucinid clams contribute to primary productivity through chemosynthesis and facilitate the growth of seagrasses (important foundation species) by lowering sulfide concentrations in tropical sediments.[52] Gammaproteobacterial symbionts of lucinid clams and stilbonematid nematodes were also recently shown to be capable of nitrogen fixation (bacterial symbiont genomes encode and express nitrogenase genes,[53] highlighting the role of symbiotic microbes in nutrient cycling in shallow marine systems.[2]
These examples demonstrate the importance of microbial symbioses for the functioning of ocean ecosystems. Understanding symbioses with this same level of detail in the context of complex communities (i.e., whole microbiomes) remains ripe for exploration and, indeed, requires a more integrated framework from the fields of microbiology, evolutionary biology, community ecology, and oceanography. Individual taxa within the microbiome may help hosts withstand a wide range of environmental conditions, including those predicted under scenarios of climate change. Next, we explore two different avenues of how interdisciplinary collaborations could advance this line of research.[2]
Examples
The microbiomes of diverse marine animals are currently under study, from simplistic organisms including sponges[55] and ctenophores [56] to more complex organisms such as sea squirts[57] and sharks.[58][1]
The relationship between the Hawaiian bobtail squid and the bioluminescent bacterium Aliivibrio fischeri is one of the best studied symbiotic relationships in the sea and is a choice system for general symbiosis research. This relationship has provided insight into fundamental processes in animal-microbial symbioses, and especially biochemical interactions and signaling between the host and bacterium.[59][60][1]
The gutless marine oligochaete worm Olavius algarvensis is another relatively well-studied marine host to microbes. These three centimetre long worms reside within shallow marine sediments of the Mediterranean Sea. The worms do not contain a mouth or a digestive or excretory system, but are instead nourished with the help of a suite of extracellular bacterial endosymbionts that reside upon coordinated use of sulfur present in the environment.[61] This system has benefited from some of the most sophisticated 'omics and visualization tools.[62] For example, multi-labeled probing has improved visualization of the microbiome[63] and transcriptomics and proteomics have been applied to examine host–microbiome interactions, including energy transfer between the host and microbes[64] and recognition of the consortia by the worm's innate immune system.[65] The major strength of this system is that it does offer the ability to study host–microbiome interactions with a low diversity microbial consortium, and it also offers a number of host and microbial genomic resources[62][66][1]
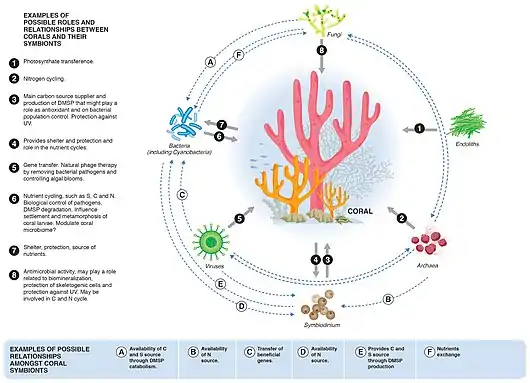
Corals
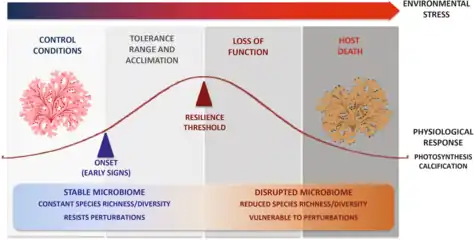
Corals are one of the more common examples of an animal host whose symbiosis with microalgae can turn to dysbiosis, and is visibly detected as bleaching. Coral microbiomes have been examined in a variety of studies, which demonstrate how variations in the ocean environment, most notably temperature, light, and inorganic nutrients, affect the abundance and performance of the microalgal symbionts, as well as calcification and physiology of the host.[68][69][1]
Studies have also suggested that resident bacteria, archaea, and fungi additionally contribute to nutrient and organic matter cycling within the coral, with viruses also possibly playing a role in structuring the composition of these members, thus providing one of the first glimpses at a multi-domain marine animal symbiosis.[70] The gammaproteobacterium Endozoicomonas is emerging as a central member of the coral's microbiome, with flexibility in its lifestyle.[71][72] Given the recent mass bleaching occurring on reefs,[73] corals will likely continue to be a useful and popular system for symbiosis and dysbiosis research.[1]
Astrangia poculata, the northern star coral, is a temperate stony coral, widely documented along the eastern coast of the United States. The coral can live with and without zooxanthellae (algal symbionts), making it an ideal model organism to study microbial community interactions associated with symbiotic state. However, the ability to develop primers and probes to more specifically target key microbial groups has been hindered by the lack of full length 16S rRNA sequences, since sequences produced by the Illumina platform are of insufficient length (approximately 250 base pairs) for the design of primers and probes.[74] In 2019, Goldsmith et al demonstrated Sanger sequencing was capable of reproducing the biologically-relevant diversity detected by deeper next-generation sequencing, while also producing longer sequences useful to the research community for probe and primer design (see diagram on right).[75]
Sponges
Sponges are common members of the ocean's diverse benthic habitats and their abundance and ability to filter large volumes of seawater have led to the awareness that these organisms play critical roles in influencing benthic and pelagic processes in the ocean.[76] They are one of the oldest lineages of animals, and have a relatively simple body plan that commonly associates with bacteria, archaea, algal protists, fungi, and viruses.[4] Sponge microbiomes are composed of specialists and generalists, and complexity of their microbiome appears to be shaped by host phylogeny.[77] Studies have shown that the sponge microbiome contributes to nitrogen cycling in the oceans, especially through the oxidation of ammonia by archaea and bacteria.[78][79] Most recently, microbial symbionts of tropical sponges were shown to produce and store polyphosphate granules,[80] perhaps enabling the host to survive periods of phosphate depletion in oligotrophic marine environments.[81] The microbiomes of some sponge species do appear to change in community structure in response to changing environmental conditions, including temperature[82] and ocean acidification,[83][84] as well as synergistic impacts.[85]

Cetaceans
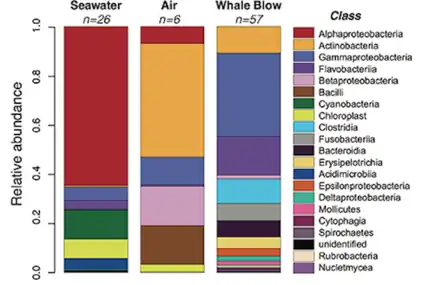
The access of microbial samples from the gut out of marine mammals is limited because most species are rare, endangered, and deep divers. There are different techniques for sampling the cetacean's gut microbiome. The most common is collecting fecal samples from the environment and taking a probe from the center that is non-contaminated.[88] Besides there are studies from rectal swabs and rare studies from stranded dead or living animals direct from the intestine.[89][90][91]
The outermost epidermal layer, i.e. the skin, is the first barrier that protects the individual from the outside world and the epidermal microbiome on it is considered an indicator not only of the health of the animal but is also considered an ecological indicator that shows the state of the surrounding environment. Knowing the microbiome of the skin of marine mammals under ''normal'' conditions has allowed us to understand how these communities are different from the free microbial communities found in the sea and how they can change according to abiotic and biotic variations, and also ''communities vary between healthy and sick individuals''.[92]
Cetaceans are in danger because they are affected by multiple stress factors which make them more vulnerable to various diseases. These animals have been noted to show high susceptibility to airway infections, but very little is known about their respiratory microbiome. Therefore, the sampling of the exhaled breath or "blow" of the cetaceans can provide an assessment of the state of health. Blow is composed of a mixture of microorganisms and organic material, including lipids, proteins , and cellular debris derived from the linings of the airways which, when released into the relatively cooler outdoor air, condense to form a visible mass of vapor, which can be collected. There are various methods for collecting exhaled breath samples, one of the most recent is through the use of aerial drones. This method provides a safer, quieter, and less invasive alternative and often a cost-effective option for monitoring fauna and flora. Once obtained, the blow samples are taken to the laboratory and we proceed with the amplification and sequencing of the respiratory tract microbiota. The use of aerial drones has been more successful with large cetaceans due to slow swim speeds and larger blow sizes.[87][93][94][95][96][86][97][98][99]
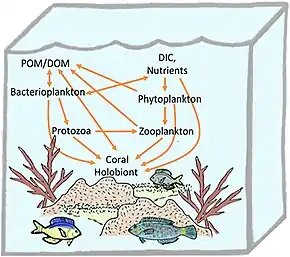
Marine holobionts
Reef-building corals are holobionts that include the coral itself (a eukaryotic invertebrate within class Anthozoa), photosynthetic dinoflagellates called zooxanthellae (Symbiodinium), and associated bacteria and viruses.[100] Co-evolutionary patterns exist for coral microbial communities and coral phylogeny.[101]
References
- Apprill, A. (2017) "Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean". Frontiers in Marine Science, 4: 222. doi:10.3389/fmars.2017.00222.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Wilkins, Laetitia G. E.; Leray, Matthieu; o'Dea, Aaron; Yuen, Benedict; Peixoto, Raquel S.; Pereira, Tiago J.; Bik, Holly M.; Coil, David A.; Duffy, J. Emmett; Herre, Edward Allen; Lessios, Harilaos A.; Lucey, Noelle M.; Mejia, Luis C.; Rasher, Douglas B.; Sharp, Koty H.; Sogin, Emilia M.; Thacker, Robert W.; Vega Thurber, Rebecca; Wcislo, William T.; Wilbanks, Elizabeth G.; Eisen, Jonathan A. (2019). "Host-associated microbiomes drive structure and function of marine ecosystems". PLOS Biology. 17 (11): e3000533. doi:10.1371/journal.pbio.3000533. PMC 6874084. PMID 31710600.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Krediet, Cory J.; Ritchie, Kim B.; Paul, Valerie J.; Teplitski, Max (2013). "Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases". Proceedings of the Royal Society B: Biological Sciences. 280 (1755). doi:10.1098/rspb.2012.2328. PMC 3574386. PMID 23363627.
- Webster, Nicole S.; Thomas, Torsten (2016). "The Sponge Hologenome". mBio. 7 (2): e00135-16. doi:10.1128/mBio.00135-16. PMC 4850255. PMID 27103626.
- Distel, D. L.; Delong, E. F.; Waterbury, J. B. (1991). "Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization". Applied and Environmental Microbiology. 57 (8): 2376–2382. doi:10.1128/AEM.57.8.2376-2382.1991. PMC 183578. PMID 1722662.
- Ruehland, Caroline; Blazejak, Anna; Lott, Christian; Loy, Alexander; Erséus, Christer; Dubilier, Nicole (2008). "Multiple bacterial symbionts in two species of co-occurring gutless oligochaete worms from Mediterranean sea grass sediments". Environmental Microbiology. 10 (12): 3404–3416. doi:10.1111/j.1462-2920.2008.01728.x. PMID 18764872.
- Nyholm, Spencer V.; McFall-Ngai, Margaret (2004). "The winnowing: Establishing the squid–vibrio symbiosis". Nature Reviews Microbiology. 2 (8): 632–642. doi:10.1038/nrmicro957. PMID 15263898. S2CID 21583331.
- Hammer, Tobin J.; Sanders, Jon G.; Fierer, Noah (2019). "Not all animals need a microbiome". FEMS Microbiology Letters. 366 (10). doi:10.1093/femsle/fnz117. PMID 31132110.
- Tipton, Laura; Darcy, John L.; Hynson, Nicole A. (2019). "A Developing Symbiosis: Enabling Cross-Talk Between Ecologists and Microbiome Scientists". Frontiers in Microbiology. 10: 292. doi:10.3389/fmicb.2019.00292. PMC 6391321. PMID 30842763.
- Apprill, Amy (2017). "Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean". Frontiers in Marine Science. 4. doi:10.3389/fmars.2017.00222. S2CID 9729436.
- McFall-Ngai, Margaret; Hadfield, Michael G.; Bosch, Thomas C. G.; Carey, Hannah V.; Domazet-Lošo, Tomislav; Douglas, Angela E.; Dubilier, Nicole; Eberl, Gerard; Fukami, Tadashi; Gilbert, Scott F.; Hentschel, Ute; King, Nicole; Kjelleberg, Staffan; Knoll, Andrew H.; Kremer, Natacha; Mazmanian, Sarkis K.; Metcalf, Jessica L.; Nealson, Kenneth; Pierce, Naomi E.; Rawls, John F.; Reid, Ann; Ruby, Edward G.; Rumpho, Mary; Sanders, Jon G.; Tautz, Diethard; Wernegreen, Jennifer J. (2013). "Animals in a bacterial world, a new imperative for the life sciences". Proceedings of the National Academy of Sciences. 110 (9): 3229–3236. Bibcode:2013PNAS..110.3229M. doi:10.1073/pnas.1218525110. PMC 3587249. PMID 23391737.
- Webster, Nicole S.; Taylor, Michael W. (2012). "Marine sponges and their microbial symbionts: Love and other relationships". Environmental Microbiology. 14 (2): 335–346. doi:10.1111/j.1462-2920.2011.02460.x. PMID 21443739.
- Azam, F.; Worden, A. Z. (2004). "OCEANOGRAPHY: Microbes, Molecules, and Marine Ecosystems". Science. 303 (5664): 1622–1624. doi:10.1126/science.1093892. PMID 15016987. S2CID 10101482.
- Cavicchioli, Ricardo; et al. (2019). "Scientists' warning to humanity: Microorganisms and climate change". Nature Reviews Microbiology. 17 (9): 569–586. doi:10.1038/s41579-019-0222-5. PMC 7136171. PMID 31213707.
- Duffy, J. Emmett; Godwin, Casey M.; Cardinale, Bradley J. (2017). "Biodiversity effects in the wild are common and as strong as key drivers of productivity". Nature. 549 (7671): 261–264. Bibcode:2017Natur.549..261D. doi:10.1038/nature23886. PMID 28869964. S2CID 4459856.
- Cardinale, Bradley J.; Duffy, J. Emmett; Gonzalez, Andrew; Hooper, David U.; Perrings, Charles; Venail, Patrick; Narwani, Anita; Mace, Georgina M.; Tilman, David; Wardle, David A.; Kinzig, Ann P.; Daily, Gretchen C.; Loreau, Michel; Grace, James B.; Larigauderie, Anne; Srivastava, Diane S.; Naeem, Shahid (2012). "Biodiversity loss and its impact on humanity" (PDF). Nature. 486 (7401): 59–67. Bibcode:2012Natur.486...59C. doi:10.1038/nature11148. PMID 22678280. S2CID 4333166.
- Gould, Alison L.; Zhang, Vivian; Lamberti, Lisa; Jones, Eric W.; Obadia, Benjamin; Korasidis, Nikolaos; Gavryushkin, Alex; Carlson, Jean M.; Beerenwinkel, Niko; Ludington, William B. (2018). "Microbiome interactions shape host fitness". Proceedings of the National Academy of Sciences. 115 (51): E11951–E11960. doi:10.1073/pnas.1809349115. PMC 6304949. PMID 30510004.
- Seemann, Janina; Yingst, Alexandra; Stuart-Smith, Rick D.; Edgar, Graham J.; Altieri, Andrew H. (2018). "The importance of sponges and mangroves in supporting fish communities on degraded coral reefs in Caribbean Panama". PeerJ. 6: e4455. doi:10.7717/peerj.4455. PMC 5878927. PMID 29610704.
- o'Brien, Paul A.; Webster, Nicole S.; Miller, David J.; Bourne, David G. (2019). "Host-Microbe Coevolution: Applying Evidence from Model Systems to Complex Marine Invertebrate Holobionts". mBio. 10 (1). doi:10.1128/mBio.02241-18. PMC 6428750. PMID 30723123.
- Croft, Martin T.; Lawrence, Andrew D.; Raux-Deery, Evelyne; Warren, Martin J.; Smith, Alison G. (2005). "Algae acquire vitamin B12 through a symbiotic relationship with bacteria". Nature. 438 (7064): 90–93. Bibcode:2005Natur.438...90C. doi:10.1038/nature04056. PMID 16267554. S2CID 4328049.
- Quigley, KM; Bay, LK; Willis, BL (2018). "Leveraging new knowledge of Symbiodinium community regulation in corals for conservation and reef restoration". Marine Ecology Progress Series. 600: 245–253. Bibcode:2018MEPS..600..245Q. doi:10.3354/meps12652. S2CID 90469901.
- Baker, David M.; Freeman, Christopher J.; Wong, Jane C.Y.; Fogel, Marilyn L.; Knowlton, Nancy (2018). "Climate change promotes parasitism in a coral symbiosis". The ISME Journal. 12 (3): 921–930. doi:10.1038/s41396-018-0046-8. PMC 5864192. PMID 29379177.
- Ngugi, David Kamanda; Miyake, Sou; Cahill, Matt; Vinu, Manikandan; Hackmann, Timothy J.; Blom, Jochen; Tietbohl, Matthew D.; Berumen, Michael L.; Stingl, Ulrich (2017). "Genomic diversification of giant enteric symbionts reflects host dietary lifestyles". Proceedings of the National Academy of Sciences. 114 (36): E7592–E7601. doi:10.1073/pnas.1703070114. PMC 5594648. PMID 28835538.
- Hoey, Andrew S.; Bellwood, David R. (2009). "Limited Functional Redundancy in a High Diversity System: Single Species Dominates Key Ecological Process on Coral Reefs". Ecosystems. 12 (8): 1316–1328. doi:10.1007/s10021-009-9291-z. S2CID 42138428.
- Nowack, Eva C. M.; Melkonian, Michael (2010). "Endosymbiotic associations within protists". Philosophical Transactions of the Royal Society B: Biological Sciences. 365 (1541): 699–712. doi:10.1098/rstb.2009.0188. PMC 2817226. PMID 20124339.
- Higgs, Nicholas D.; Newton, Jason; Attrill, Martin J. (2016). "Caribbean Spiny Lobster Fishery is Underpinned by Trophic Subsidies from Chemosynthetic Primary Production". Current Biology. 26 (24): 3393–3398. doi:10.1016/j.cub.2016.10.034. hdl:10026.1/9129. PMID 27939312. S2CID 14401680.
- Seah, Brandon K. B.; Antony, Chakkiath Paul; Huettel, Bruno; Zarzycki, Jan; Schada von Borzyskowski, Lennart; Erb, Tobias J.; Kouris, Angela; Kleiner, Manuel; Liebeke, Manuel; Dubilier, Nicole; Gruber-Vodicka, Harald R. (2019). "Sulfur-Oxidizing Symbionts without Canonical Genes for Autotrophic CO2Fixation". mBio. 10 (3). doi:10.1128/mBio.01112-19. PMC 6593406. PMID 31239380.
- Gast, Rebecca J.; Sanders, Robert W.; Caron, David A. (2009). "Ecological strategies of protists and their symbiotic relationships with prokaryotic microbes". Trends in Microbiology. 17 (12): 563–569. doi:10.1016/j.tim.2009.09.001. PMID 19828317.
- Huang, Ying; Callahan, Sean; Hadfield, Michael G. (2012). "Recruitment in the sea: Bacterial genes required for inducing larval settlement in a polychaete worm". Scientific Reports. 2: 228. Bibcode:2012NatSR...2E.228H. doi:10.1038/srep00228. PMC 3260340. PMID 22355742. S2CID 14731587.
- Delannoy, Sabine; Chaves, Byron D.; Ison, Sarah A.; Webb, Hattie E.; Beutin, Lothar; Delaval, José; Billet, Isabelle; Fach, Patrick (2016). "Revisiting the STEC Testing Approach: Using espK and espV to Make Enterohemorrhagic Escherichia coli (EHEC) Detection More Reliable in Beef". Frontiers in Microbiology. 7: 1. doi:10.3389/fmicb.2016.00001. PMC 4722105. PMID 26834723.
- Wheeler, Glen L.; Tait, Karen; Taylor, Alison; Brownlee, Colin; Joint, IAN (2006). "Acyl-homoserine lactones modulate the settlement rate of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism". Plant, Cell and Environment. 29 (4): 608–618. doi:10.1111/j.1365-3040.2005.01440.x. PMID 17080611.
- Provasoli, Luigi; Pintner, Irma J. (1980). "Bacteria Induced Polymorphism in an Axenic Laboratory Strain of Ulva Lactuca (Chlorophyceae)1". Journal of Phycology. 16 (2): 196–201. doi:10.1111/j.1529-8817.1980.tb03019.x. S2CID 85817449.
- Matsuo, Y.; Imagawa, H.; Nishizawa, M.; Shizuri, Y. (2005). "Isolation of an Algal Morphogenesis Inducer from a Marine Bacterium". Science. 307 (5715): 1598. doi:10.1126/science.1105486. PMID 15761147. S2CID 28850526.
- Goecke, F.; Labes, A.; Wiese, J.; Imhoff, JF (2010). "Chemical interactions between marine macroalgae and bacteria". Marine Ecology Progress Series. 409: 267–299. Bibcode:2010MEPS..409..267G. doi:10.3354/meps08607.
- Amin, S. A.; Hmelo, L. R.; Van Tol, H. M.; Durham, B. P.; Carlson, L. T.; Heal, K. R.; Morales, R. L.; Berthiaume, C. T.; Parker, M. S.; Djunaedi, B.; Ingalls, A. E.; Parsek, M. R.; Moran, M. A.; Armbrust, E. V. (2015). "Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria". Nature. 522 (7554): 98–101. Bibcode:2015Natur.522...98A. doi:10.1038/nature14488. PMID 26017307. S2CID 4462055.
- Celdrán, D.; Espinosa, E.; Sánchez-Amat, A.; Marín, A. (2012). "Effects of epibiotic bacteria on leaf growth and epiphytes of the seagrass Posidonia oceanica". Marine Ecology Progress Series. 456: 21–27. Bibcode:2012MEPS..456...21C. doi:10.3354/meps09672.
- Woznica, Arielle; King, Nicole (2018). "Lessons from simple marine models on the bacterial regulation of eukaryotic development". Current Opinion in Microbiology. 43: 108–116. doi:10.1016/j.mib.2017.12.013. PMC 6051772. PMID 29331767.
- McFall-Ngai, Margaret J. (2014). "The Importance of Microbes in Animal Development: Lessons from the Squid-Vibrio Symbiosis". Annual Review of Microbiology. 68: 177–194. doi:10.1146/annurev-micro-091313-103654. PMC 6281398. PMID 24995875.
- Shikuma, Nicholas J.; Antoshechkin, Igor; Medeiros, João M.; Pilhofer, Martin; Newman, Dianne K. (2016). "Stepwise metamorphosis of the tubeworm Hydroides elegansis mediated by a bacterial inducer and MAPK signaling". Proceedings of the National Academy of Sciences. 113 (36): 10097–10102. doi:10.1073/pnas.1603142113. PMC 5018781. PMID 27551098. S2CID 23501584.
- Krediet, Cory J.; Carpinone, Emily M.; Ritchie, Kim B.; Teplitski, Max (2013). "Characterization of thegacA-dependent surface and coral mucus colonization by an opportunistic coral pathogen Serratia marcescensPDL100". FEMS Microbiology Ecology. 84 (2): 290–301. doi:10.1111/1574-6941.12064. PMID 23278392.
- Pita L, Rix L, Slaby BM, Franke A, Hentschel U (March 2018). "The sponge holobiont in a changing ocean: from microbes to ecosystems". Microbiome. 6 (1): 46. doi:10.1186/s40168-018-0428-1. PMC 5845141. PMID 29523192.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Steinberg, Peter D.; De Nys, Rocky (2002). "Chemical Mediation of Colonization of Seaweed Surfaces1". Journal of Phycology. 38 (4): 621–629. doi:10.1046/j.1529-8817.2002.02042.x. S2CID 83963124.
- Dobretsov, Sergey; Abed, Raeid M.M.; Teplitski, Max (2013). "Mini-review: Inhibition of biofouling by marine microorganisms". Biofouling. 29 (4): 423–441. doi:10.1080/08927014.2013.776042. PMID 23574279. S2CID 34459128.
- Reed, Daniel C.; Carlson, Craig A.; Halewood, Elisa R.; Nelson, J. Clinton; Harrer, Shannon L.; Rassweiler, Andrew; Miller, Robert J. (2015). "Patterns and controls of reef-scale production of dissolved organic carbon by giant kelp M acrocystis pyrifera". Limnology and Oceanography. 60 (6): 1996–2008. Bibcode:2015LimOc..60.1996R. doi:10.1002/lno.10154. S2CID 85962482.
- Vollmers, John; Wiegand, Sandra; Kaster, Anne-Kristin (2017). "Comparing and Evaluating Metagenome Assembly Tools from a Microbiologist's Perspective - Not Only Size Matters!". PLOS ONE. 12 (1): e0169662. Bibcode:2017PLoSO..1269662V. doi:10.1371/journal.pone.0169662. PMC 5242441. PMID 28099457.
- Elshahawi, Sherif I.; Trindade-Silva, Amaro E.; Hanora, Amro; Han, Andrew W.; Flores, Malem S.; Vizzoni, Vinicius; Schrago, Carlos G.; Soares, Carlos A.; Concepcion, Gisela P.; Distel, Dan L.; Schmidt, Eric W.; Haygood, Margo G. (2013). "Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills". Proceedings of the National Academy of Sciences. 110 (4): E295–E304. doi:10.1073/pnas.1213892110. PMC 3557025. PMID 23288898.
- De Goeij, Jasper M.; Van Oevelen, Dick; Vermeij, Mark J. A.; Osinga, Ronald; Middelburg, Jack J.; De Goeij, Anton F. P. M.; Admiraal, Wim (2013). "Surviving in a Marine Desert: The Sponge Loop Retains Resources within Coral Reefs". Science. 342 (6154): 108–110. Bibcode:2013Sci...342..108D. doi:10.1126/science.1241981. PMID 24092742. S2CID 6720678.
- Colman, Albert S. (2015). "Sponge symbionts and the marine P cycle". Proceedings of the National Academy of Sciences. 112 (14): 4191–4192. Bibcode:2015PNAS..112.4191C. doi:10.1073/pnas.1502763112. PMC 4394276. PMID 25825737.
- Corzett, Christopher H.; Elsherbini, Joseph; Chien, Diana M.; Hehemann, Jan-Hendrik; Henschel, Andreas; Preheim, Sarah P.; Yu, Xiaoqian; Alm, Eric J.; Polz, Martin F. (2018). "Evolution of a Vegetarian Vibrio: Metabolic Specialization of Vibrio breoganii to Macroalgal Substrates". Journal of Bacteriology. 200 (15). doi:10.1128/JB.00020-18. PMC 6040190. PMID 29632094.
- Bengtsson, MM; Sjøtun, K.; Storesund, JE; Øvreås, J. (2011). "Utilization of kelp-derived carbon sources by kelp surface-associated bacteria". Aquatic Microbial Ecology. 62 (2): 191–199. doi:10.3354/ame01477. hdl:1956/4610.
- Pfister, Catherine A.; Altabet, Mark A.; Weigel, Brooke L. (2019). "Kelp beds and their local effects on seawater chemistry, productivity, and microbial communities". Ecology. 100 (10): e02798. doi:10.1002/ecy.2798. PMID 31233610. S2CID 195355739.
- Van Der Heide, T.; Govers, L. L.; De Fouw, J.; Olff, H.; Van Der Geest, M.; Van Katwijk, M. M.; Piersma, T.; Van De Koppel, J.; Silliman, B. R.; Smolders, A. J. P.; Van Gils, J. A. (2012). "A Three-Stage Symbiosis Forms the Foundation of Seagrass Ecosystems". Science. 336 (6087): 1432–1434. Bibcode:2012Sci...336.1432V. doi:10.1126/science.1219973. hdl:11370/23625acb-7ec0-4480-98d7-fad737d7d4fe. PMID 22700927. S2CID 27806510.
- Petersen, Jillian M.; Kemper, Anna; Gruber-Vodicka, Harald; Cardini, Ulisse; Van Der Geest, Matthijs; Kleiner, Manuel; Bulgheresi, Silvia; Mußmann, Marc; Herbold, Craig; Seah, Brandon K.B.; Antony, Chakkiath Paul; Liu, Dan; Belitz, Alexandra; Weber, Miriam (2017). "Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation". Nature Microbiology. 2 (1): 16195. doi:10.1038/nmicrobiol.2016.195. PMC 6872982. PMID 27775707.
- Peixoto, R.S., Rosado, P.M., Leite, D.C.D.A., Rosado, A.S. and Bourne, D.G. (2017) "Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience". Frontiers in Microbiology, 8: 341. doi:10.3389/fmicb.2017.00341.
- Webster, N.S., Negri, A.P., Botté, E.S., Laffy, P.W., Flores, F., Noonan, S., Schmidt, C. and Uthicke, S. (2016) "Host-associated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification". Scientific reports, 6: 19324. doi:10.1038/srep19324.
- Daniels, C. and Breitbart, M. (2012) "Bacterial communities associated with the ctenophores Mnemiopsis leidyi and Beroe ovata". FEMS Microbiology Ecology, 82(1): 90–101. doi:10.1111/j.1574-6941.2012.01409.x.
- Blasiak, L.C., Zinder, S.H., Buckley, D.H. and Hill, R.T. (2014) "Bacterial diversity associated with the tunic of the model chordate Ciona intestinalis". The ISME Journal, 8(2): 309–320. doi:10.1038/ismej.2013.156.
- Givens, C.E., Ransom, B., Bano, N. and Hollibaugh, J.T. (2015) "Comparison of the gut microbiomes of 12 bony fish and 3 shark species". Marine Ecology Progress Series, 518: 209–223. doi:10.3354/meps11034.
- McFall-Ngai, M.J. (2000) "Negotiations between animals and bacteria: the 'diplomacy'of the squid-vibrio symbiosis". Comparative Biochemistry and Physiology, Part A: Molecular & Integrative Physiology, 126(4): 471–480. doi:10.1016/S1095-6433(00)00233-6.
- McFall-Ngai, M. (2014) "Divining the essence of symbiosis: insights from the squid-vibrio model". PLoS Biology, 12(2): e1001783. doi:10.1371/journal.pbio.1001783.
- Dubilier, N., Mülders, C., Ferdelman, T., de Beer, D., Pernthaler, A., Klein, M., Wagner, M., Erséus, C., Thiermann, F., Krieger, J. and Giere, O. (2001) "Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm". Nature, 411(6835): 298–302. doi:10.1038/35077067.
- Woyke, T., Teeling, H., Ivanova, N.N., Huntemann, M., Richter, M., Gloeckner, F.O., Boffelli, D., Anderson, I.J., Barry, K.W., Shapiro, H.J. and Szeto, E. (2006) "Symbiosis insights through metagenomic analysis of a microbial consortium". Nature, 443(7114): 950–955. doi:10.1038/nature05192.
- Schimak, M.P., Kleiner, M., Wetzel, S., Liebeke, M., Dubilier, N. and Fuchs, B.M. (2016) "MiL-FISH: Multilabeled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells". Applied and Environmental Microbiology, 82(1): 62–70. doi:10.1128/AEM.02776-15.
- Kleiner, M., Wentrup, C., Lott, C., Teeling, H., Wetzel, S., Young, J., Chang, Y.J., Shah, M., VerBerkmoes, N.C., Zarzycki, J. and Fuchs, G. (2012) "Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use". Proceedings of the National Academy of Sciences, 109(19): E1173–E1182. doi:10.1073/pnas.1121198109.
- Wippler, J., Kleiner, M., Lott, C., Gruhl, A., Abraham, P.E., Giannone, R.J., Young, J.C., Hettich, R.L. and Dubilier, N. (2016) "Transcriptomic and proteomic insights into innate immunity and adaptations to a symbiotic lifestyle in the gutless marine worm Olavius algarvensis". BMC Genomics, 17(1): 942. doi:10.1186/s12864-016-3293-y.
- Ruehland, C., Blazejak, A., Lott, C., Loy, A., Erséus, C. and Dubilier, N. (2008) "Multiple bacterial symbionts in two species of co‐occurring gutless oligochaete worms from Mediterranean sea grass sediments". Environmental microbiology, 10(12): 3404–3416. doi:10.1111/j.1462-2920.2008.01728.x.
- Sharp, Koty H.; Pratte, Zoe A.; Kerwin, Allison H.; Rotjan, Randi D.; Stewart, Frank J. (2017). "Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata". Microbiome. 5 (1): 120. doi:10.1186/s40168-017-0329-8. PMC 5603060. PMID 28915923.
- Dubinsky, Z. and Jokiel, P.L. (1994) "Ratio of energy and nutrient fluxes regulates symbiosis between zooxanthellae and corals". Pacific Science, 48(3): 313–324.
- Anthony, K.R., Kline, D.I., Diaz-Pulido, G., Dove, S. and Hoegh-Guldberg, O.(2008) "Ocean acidification causes bleaching and productivity loss in coral reef builders". Proceedings of the National Academy of Sciences, 105(45): 17442–17446. doi:10.1073/pnas.0804478105.
- Bourne, D.G., Morrow, K.M. and Webster, N.S. (2016) "Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems". Annual Review of Microbiology, 70: 317–340. doi:10.1146/annurev-micro-102215-095440.
- Neave, M.J., Apprill, A., Ferrier-Pagès, C. and Voolstra, C.R. (2016) "Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas". Applied Microbiology and Biotechnology, 100(19): 8315–8324. doi:10.1007/s00253-016-7777-0.
- Neave, M.J., Michell, C.T., Apprill, A. and Voolstra, C.R. (2017) "Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts". Scientific Reports, 7: 40579. doi:10.1038/srep40579.
- Hughes, T.P., Kerry, J.T., Álvarez-Noriega, M., Álvarez-Romero, J.G., Anderson, K.D., Baird, A.H., Babcock, R.C., Beger, M., Bellwood, D.R., Berkelmans, R. and Bridge, T.C. (2017) "Global warming and recurrent mass bleaching of corals". Nature, 543(7645): 373–377. doi:10.1038/nature21707.
- USGS scientists publish long-read microbiome sequences from temperate coral, providing community resource for probe and primer design, United States Geological Survey, 6 March 2019.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - b. Goldsmith, Dawn; a. Pratte, Zoe; a. Kellogg, Christina; e. Snader, Sara; h. Sharp, Koty (2019). "Stability of temperate coral Astrangia poculata microbiome is reflected across different sequencing methodologies". AIMS Microbiology. 5 (1): 62–76. doi:10.3934/microbiol.2019.1.62. PMC 6646935. PMID 31384703.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Bell, J.J. (2008) "The functional roles of marine sponges". Estuarine, Coastal and Shelf Science, 79(3): 341–353. doi:10.1016/j.ecss.2008.05.002.
- Thomas, T., Moitinho-Silva, L., Lurgi, M., Björk, J.R., Easson, C., Astudillo-García, C., Olson, J.B., Erwin, P.M., López-Legentil, S., Luter, H. and Chaves-Fonnegra, A. (2016) "Diversity, structure and convergent evolution of the global sponge microbiome". Nature Communications, 7(1): 1-12. doi:10.1038/ncomms11870.
- Bayer, K., Schmitt, S. and Hentschel, U. (2008) "Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba". Environmental Microbiology, 10(11): 2942–2955. doi:10.1111/j.1462-2920.2008.01582.x.
- Radax, R., Hoffmann, F., Rapp, H.T., Leininger, S. and Schleper, C. (2012) "Ammonia‐oxidizing archaea as main drivers of nitrification in cold‐water sponges". Environmental Microbiology, 14(4): 909_923. doi:10.1111/j.1462-2920.2011.02661.x.
- Zhang, F., Blasiak, L.C., Karolin, J.O., Powell, R.J., Geddes, C.D. and Hill, R.T. (2015) "Phosphorus sequestration in the form of polyphosphate by microbial symbionts in marine sponges". Proceedings of the National Academy of Sciences, 112(14): 4381–4386. doi:10.1073/pnas.1423768112.
- Colman, A.S. (2015) "Sponge symbionts and the marine P cycle". Proceedings of the National Academy of Sciences, 112(14): 4191–4192. doi:10.1073/pnas.1502763112.
- Simister, R., Taylor, M.W., Tsai, P., Fan, L., Bruxner, T.J., Crowe, M.L. and Webster, N. (2012) "Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environmental Microbiology, 14(12): 3232–3246. doi:10.1111/1462-2920.12010.
- Morrow, K.M., Bourne, D.G., Humphrey, C., Botté, E.S., Laffy, P., Zaneveld, J., Uthicke, S., Fabricius, K.E. and Webster, N.S. (2015) "Natural volcanic CO 2 seeps reveal future trajectories for host–microbial associations in corals and sponges". The ISME Journal, 9(4): 894–908. doi:10.1038/ismej.2014.188.
- Ribes, M., Calvo, E., Movilla, J., Logares, R., Coma, R. and Pelejero, C. (2016) "Restructuring of the sponge microbiome favors tolerance to ocean acidification. Environmental Microbiology Reports, 8(4): 536–544. doi:10.1111/1758-2229.12430.
- Lesser, M.P., Fiore, C., Slattery, M. and Zaneveld, J. (2016) "Climate change stressors destabilize the microbiome of the Caribbean barrel sponge, Xestospongia muta". Journal of Experimental Marine Biology and Ecology, 475: 11–18. doi:10.1016/j.jembe.2015.11.004.
- Acevedo-Whitehouse K, Rocha-Gosselin A, Gendron D (April 2010). "A novel non‐invasive tool for disease surveillance of free‐ranging whales and its relevance to conservation programs". Animal Conservation. 13 (2): 217–225. doi:10.1111/j.1469-1795.2009.00326.x. S2CID 86518859.
- Pirotta V, Smith A, Ostrowski M, Russell D, Jonsen ID, Grech A, Harcourt R (December 2017). "An economical custom-built drone for assessing whale health". Frontiers in Marine Science. 4: 425. doi:10.3389/fmars.2017.00425.
- Suzuki A, Ueda K, Segawa T, Suzuki M (June 2019). "Fecal microbiota of captive Antillean manatee Trichechus manatus manatus". FEMS Microbiology Letters. 366 (11). doi:10.1093/femsle/fnz134. PMID 31210263.
- Sehnal L, Brammer-Robbins E, Wormington AM, Blaha L, Bisesi J, Larkin I, et al. (2021). "Microbiome Composition and Function in Aquatic Vertebrates: Small Organisms Making Big Impacts on Aquatic Animal Health". Frontiers in Microbiology. 12: 567408. doi:10.3389/fmicb.2021.567408. PMC 7995652. PMID 33776947.
- Bik EM, Costello EK, Switzer AD, Callahan BJ, Holmes SP, Wells RS, et al. (February 2016). "Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea". Nature Communications. 7: 10516. Bibcode:2016NatCo...710516B. doi:10.1038/ncomms10516. PMC 4742810. PMID 26839246.
- Bai S, Zhang P, Lin M, Lin W, Yang Z, Li S (May 2021). "Microbial diversity and structure in the gastrointestinal tracts of two stranded short-finned pilot whales (Globicephala macrorhynchus) and a pygmy sperm whale (Kogia breviceps)". Integrative Zoology. 16 (3): 324–335. doi:10.1111/1749-4877.12502. PMC 9292824. PMID 33174288. S2CID 226302293.
- Apprill A, Mooney TA, Lyman E, Stimpert AK, Rappé MS (April 2011). "Humpback whales harbour a combination of specific and variable skin bacteria". Environmental Microbiology Reports. 3 (2): 223–232. doi:10.1111/j.1758-2229.2010.00213.x. PMID 23761254.
- Apprill A, Miller CA, Moore MJ, Durban JW, Fearnbach H, Barrett-Lennard LG (2017). "Extensive Core Microbiome in Drone-Captured Whale Blow Supports a Framework for Health Monitoring". mSystems. 2 (5). doi:10.1128/mSystems.00119-17. PMC 5634792. PMID 29034331.
- Vendl C, Ferrari BC, Thomas T, Slavich E, Zhang E, Nelson T, Rogers T (August 2019). "Interannual comparison of core taxa and community composition of the blow microbiota from East Australian humpback whales". FEMS Microbiology Ecology. 95 (8). doi:10.1093/femsec/fiz102. PMID 31260051.
- Johnson WR, Torralba M, Fair PA, Bossart GD, Nelson KE, Morris PJ (December 2009). "Novel diversity of bacterial communities associated with bottlenose dolphin upper respiratory tracts". Environmental Microbiology Reports. 1 (6): 555–62. doi:10.1111/j.1758-2229.2009.00080.x. PMID 23765934.
- Centelleghe C, Carraro L, Gonzalvo J, Rosso M, Esposti E, Gili C, Bonato M, Pedrotti D, Cardazzo B, Povinelli M, Mazzariol S (2020). "The use of Unmanned Aerial Vehicles (UAVs) to sample the blow microbiome of small cetaceans". PLOS ONE. 15 (7): e0235537. Bibcode:2020PLoSO..1535537C. doi:10.1371/journal.pone.0235537. PMC 7332044. PMID 32614926.
- Raverty SA, Rhodes LD, Zabek E, Eshghi A, Cameron CE, Hanson MB, Schroeder JP (March 2017). "Respiratory Microbiome of Endangered Southern Resident Killer Whales and Microbiota of Surrounding Sea Surface Microlayer in the Eastern North Pacific". Scientific Reports. 7 (1): 394. Bibcode:2017NatSR...7..394R. doi:10.1038/s41598-017-00457-5. PMC 5428453. PMID 28341851.
- Lima N, Rogers T, Acevedo-Whitehouse K, Brown MV (February 2012). "Temporal stability and species specificity in bacteria associated with the bottlenose dolphins respiratory system". Environmental Microbiology Reports. 4 (1): 89–96. doi:10.1111/j.1758-2229.2011.00306.x. PMID 23757234.
- Geoghegan JL, Pirotta V, Harvey E, Smith A, Buchmann JP, Ostrowski M, Eden JS, Harcourt R, Holmes EC (June 2018). "Virological Sampling of Inaccessible Wildlife with Drones". Viruses. 10 (6): 300. doi:10.3390/v10060300. PMC 6024715. PMID 29865228.
- Knowlton N, Rohwer F (October 2003). "Multispecies microbial mutualisms on coral reefs: the host as a habitat". The American Naturalist. 162 (4 Suppl): S51–62. doi:10.1086/378684. PMID 14583857. S2CID 24127308.
- Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, et al. (November 2018). "Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny". Nature Communications. 9 (1): 4921. doi:10.1038/s41467-018-07275-x. PMC 6250698. PMID 30467310.
- Thompson JR, Rivera HE, Closek CJ, Medina M (2014). "Microbes in the coral holobiont: partners through evolution, development, and ecological interactions". Frontiers in Cellular and Infection Microbiology. 4: 176. doi:10.3389/fcimb.2014.00176. PMC 4286716. PMID 25621279.
- Ugarelli K, Chakrabarti S, Laas P, Stingl U (December 2017). "The Seagrass Holobiont and Its Microbiome". Microorganisms. 5 (4): 81. doi:10.3390/microorganisms5040081. PMC 5748590. PMID 29244764.
 Modified text was copied from this source, which is available under a [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License
Modified text was copied from this source, which is available under a [https://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International License - Cavalcanti GS, Shukla P, Morris M, Ribeiro B, Foley M, Doane MP, Thompson CC, Edwards MS, Dinsdale EA, Thompson FL (September 2018). "Rhodoliths holobionts in a changing ocean: host-microbes interactions mediate coralline algae resilience under ocean acidification". BMC Genomics. 19 (1): 701. doi:10.1186/s12864-018-5064-4. PMC 6154897. PMID 30249182.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Further references
- Stal, L. J. and Cretoiu, M. S. (Eds.) (2016) The marine microbiome: an untapped source of biodiversity and biotechnological potential Springer. ISBN 9783319330006.
- Marine Microbiome and Biogeochemical Cycles in Marine Productive Areas. Frontiers Media S.A. 2020. ISBN 978-2-88963-276-3. OCLC 1291256407.