Multiple Epidermal Growth Factor-like Domains 8
Megf8 also known as Multiple Epidermal Growth Factor-like Domains 8, is a protein coding gene that encodes a single pass membrane protein, known to participate in developmental regulation and cellular communication.[5] It is located on chromosome 19 at the 49th open reading frame in humans (19q13.2).[6] There are two isoform constructs known for MEGF8, which differ by a 67 amino acid indel. The isoform 2 splice version (analyzed throughout this page) is 2785 amino acids long, and predicted to be 296.6 kdal in mass. Isoform 1 is composed of 2845 amino acids and predicted to weigh 303.1 kdal. Using BLAST searches, orthologs were found primarily in mammals, but MEGF8 is also conserved in invertebrates and fishes, and rarely in birds, reptiles, and amphibians. A notably important paralog to multiple epidermal growth factor-like domains 8 is ATRNL1 (Attractin-like 1), which is also a single pass transmembrane protein, with several of the same key features and motifs as MEGF8, as indicated by Simple Modular Architecture Research Tool[7] (SMART) which is hosted by the European Molecular Biology Laboratory located in Heidelberg, Germany. MEGF8 has been predicted to be a key player in several developmental processes, such as left-right patterning and limb formation. Currently, researchers have found MEGF8 SNP mutations to be the cause of Carpenter syndrome subtype 2.
Gene
Evolution & Orthologs
A fairly highly conserved protein, MEGF8 has conserved orthologs from P. paniscus to N. vectensis. Orthologs are found in mammals, amphibians, fish, insects, crustaceans, and invertebrates.[8] Organization of the data showed that as time since divergence between humans and orthologs increased, the sequence identity decreased.
| Genus/Species | Organism Common Name | Accession Number | Sequence Identity | Sequence Similarity | Length (AAs) |
| Pan Paniscus | Pygmy Chimpanzee | XP_003811808 | 99% | 99% | 2778 |
| Bos Mutus | Yak | XP_005909034 | 79% | 82% | 2842 |
| Orcinius Orca | Orca Whale | XP_004271289 | 93% | 94% | 2789 |
| Trichechus manatus latirostris | Florida Manatee | XP_004388865 | 88% | 89% | 2708 |
| Leptonychotes weddellii | Weddell Seal | XP_006748348 | 91% | 92% | 2068 |
| Rattus norvegicus | Rat | NP_446080.1 | 88% | 89% | 2789 |
| Mus musculus | Mouse | NP_001153872.1 | 89% | 90% | 2789 |
| Ophiophagus hannah | King Cobra | ETE71721 | 63% | 70% | 404 |
| Alligator mississippiensis | American Alligator | XP_006273703 | 63% | 71% | 2793 |
| Alligator sinensis | Chinese alligator | XP_006038171 | 67% | 75% | 2465 |
| Xenopus tropicalis | Western clawed frog | XP_002936442 | 56% | 67% | 2730 |
| Neolamprologus brichardi | African Cichlid | XP_006808273 | 55% | 67% | 2813 |
| Danio rerio | Zebra fish | XP_005158088 | 54% | 66% | 2870 |
| IIctalurus punctatus | Channel Catfish | AHI50432 | 54% | 77% | 2875 |
| Oryzias latipes | Japanese Rice Fish | XP_004078282 | 54% | 67% | 2952 |
| Apis mellifera | Western Honey Bee | XP_006568067 | 31% | 45% | 2913 |
| Ceratitis capitata | Mediterranean Fruit Fly | JAB95791 | 32% | 45% | 2959 |
| Daphnia pulex | Common Water Flea | EFX84934 | 35% | 48% | 2888 |
| Strongylocentrotus purpuratus | Purple Sea Urchin | XP_789561 | 37% | 51% | 194 |
| Nematostella vectensis | Starlet Sea Anemone | XP_001635521 | 38% | 51% | 2534 |
Paralogs
MEGF8 has one known paralog: ATRNL1. The ATRNL1 protein is approximately half the length of MEGF8, and contains several of the same conserved domains, including the CUB domain and transmembrane sequence. It is key to note that ATRNL1 is found in many birds and amphibians, where MEGF8 is not found in any birds, and only one amphibian.
Promoters
Genomatix's ElDorado (http://www.genomatix.de/), a gene promoter database, predicted ten different possible promoters for megf8. The promoter having promoter ID number GXP_1262882 and transcript ID GXT_22531930, was predicted with the highest confidence. This promoter is located on the plus strand of chromosome 19, ranging from nucleotide 42829077 to 42830497, making it a 1421 nucleotide long sequence. The promoter sequence overlaps with the transcriptional start codon in the gene.
Transcription Factors
More than one hundred transcription factor binding sites were predicted to be found in the megf8 promoter region through Genomatix. The top twenty most confidently predicted factors include the following:
- Ccaat/Enhancer Binding Protein
- Vertebrate TATA binding protein factor
- CCAAT binding factors
- Activator-, mediator- and TBP-dependent core promoter element for RNA polymerase II transcription from TATA-less promoters
- Chorion-specific transcription factors with a GCM DNA binding domain
- Signal transducer and activator of transcription
- Heat shock factors (2 sites)
- GC-Box factors SP1/GC
- Estrogen response elements
- KRAB domain zinc finger protein 57
- Neuron-specific olfactory factor (2 sites)
- Nuclear respiratory factor 1
- RXR heterodimer binding sites
- GATA binding factors
- Nuclear receptor subfamily 2 factors
- Octamer binding protein
- EGR/nerve growth factor induced protein C & related factors
- Neuron-restrictive silencer factor
Protein Architecture
Primary Structure
MEGF8 is composed of either 2845 amino acids (Isoform 1) or 2778 amino acids (Isoform 2). Isoform 2 undergoes a 67 amino acid removal from 700-766, which accounts for its shortened length; otherwise, the two isoforms are identical. Using SAPS, a Statistical Analysis of Protein Sequence [9] software, amino acid bias was able to be determined. Isoform one is rich in cysteine and glycine, and deficient in isoleucine and lysine. Isoform 2 of MEGF8 was found to have very high levels of cysteine, moderately high levels of glycine, and low levels of isoleucine and lysine. The high levels of cysteine residues contributes to the numerous disulfide bonds found in the mature protein's folded structure. Overall, MEGF8 has a pH between 6.4 and 7.0, depending on the organism's sequence. Human MEGF8's pH is 6.4. This nearly neutral pH enables the protein to fold properly and inhibits denaturation. The twenty most conserved amino acids, found through a multiple sequence alignment of 20 orthologs, were found to be located in the CUB and transmembrane domains.
Secondary Structure
Prediction software PELE[10] from UCSC Biology Workbench indicated that MEGF8 is primarily composed of beta-folded sheets, with occasional short alpha helix segments. PELE uses eight different prediction programs to compare and confirm predictions, enhancing the confidence level. The beta-folded sheets occur at many of the key domains, including the EGF-domains, kelch domains, and EGF-laminin domains. This information from PELE also corresponded with the secondary structure and 3D structure predictions made by PHYRE2[11]
Predicted Key Domains & Features
MEGF8 is predicted to contain several different types of features, domains, and motifs that play a key role in the protein's function, structure, and location. These are listed in Table 1. Functions, found through SMART[7] analysis, as well as NCBI Conserved Domains Search[12] include:
- CUB domain: extracellular domain: present in proteins mostly known to be involved in development.
- Epidermal Growth Factor Domain: a short peptide with a distinctive motif of six cysteines, which is found in many different proteins of diverse functions[13]
- EGF-like domain: contains several sub-families of different functions according to location and protein; not specified for MEGF8.
- Calcium EGF-like domain: Calcium-binding EGF-like domain, present in a large number of membrane-bound and extracellular (mostly animal) proteins. Many of these proteins require calcium for their biological function and calcium-binding sites have been found to be located at the N-terminus of particular EGF-like domains.
- Kelch motif: Galactose oxidase, central domain; Found to cause formation of ß propeller tertiary structure of the protein.
- Leucine Zipper: A motif found in regulatory proteins, as predicted by PSORT II[14]
- Laminin EGF-like domain: laminins are the major noncollagenous components of basement membranes that mediate cell adhesion, growth migration, and differentiation; the laminin-type epidermal growth factor-like module occurs in tandem arrays; the domain contains 4 disulfide bonds (loops a-d) the first three resemble epidermal growth factor (EGF).
- PSI domain: domain found in plexins, semaphorins and integrins. Plexin are involved in the development of neural and epithelial tissues; semaphorins induce the collapse and paralysis of neuronal growth cones; and integrins may mediate adhesive or migratory functions of epithelial cells.
Predicted Domain & Motif Locations
| Feature, Domain, or Motif Name | Number in MEGF8 | Amino Acid Location Range (1-2785) |
| Signal Peptide | 1 | 1-34 |
| CUB Domain | 1 | 40-147 |
| Epidermal Growth Factor (EGF) Domain | 6 | 148-177; 180-210; 1057-1100; 2121-2160; 2162-2190; 2200-2240 |
| D1k3ia Structural Domain | 2 | 233-550; 1449-1801 |
| Kelch Repeat | 9 | 241-276; 340-388; 454-504; 519-575; 1450-1492; 1505-1552; 1724-1764; 1780-1820; 2239-2255 |
| Leucine Zipper Pattern | 1 | 1698-1719 |
| PSI Domain | 6 | 787-839; 889-931; 945-1013; 1864-1919; 2008-2058; 2060-2117 |
| EGF_Ca Domain | 1 | 1014-1055 |
| EGF_Like Domain | 4 | 1103-1148; 1346-1485; 2244-2317; 2320-2381 |
| EGF_LAM Domain | 1 | 1151-1199 |
| Transmembrane Region | 1 | 2588-2610 |


Tertiary Structure
One of the key attributes of MEGF8's tertiary structure is its 7-bladed beta propeller which is formed by the kelch motif found in its D1k3ia3 structural domain, which was identified by SCOP. SCOP[15] also indicated that the beta-propeller in MEGF8 is a member of the galactose oxidase super family. Each of the seven blades are made up of a four stranded beta-folded motifs. It is also important to note that although many phosphorylation sites are predicted at high confidence, several other topographic predictions (i.e. disulfide bonds, glycosylation, other extracellular features), do not support these predictions.
Predicted Post Translational Modifications
| Feature | Number Predicted in MEGF8 | Amino Acid Location Range (1-2785) | Source |
| Cysteine involved in Disulfide Bond | 99+ Possible Sites | - | DISULFIND[16] & UniProt |
| SUMOylation | 3 (confidently) | K886; K1681; K1737 | SUMOplot[17] |
| Phosphorylation | 116 | - | NetPhos[18] |
| Internal Repeats | 1 | CQCNGH 1144-1149 & 2313-2318 | SAPS[19] |
| N-linked Glycosylation | 20 | 56; 223; 267; 427; 699; 749; 968; 987; 1054; 1140; 1210; 1539; 1908; 1929; 2006; 2153; 2168; 2340; 2778 | NetNGlyc[20] |
| Signal Peptide Cleavage | 1 | between amino acids 34 and 35 | SignalP[21] |
| Hydrophobic Domain | 1 | 2588-2610 | SAPS |
| Extracellular Domain | 1 | 1- 2587 | Phobius[22] |
| Transmembrane Region | 1 | 2588-2610 | Phobius, SAPS, SMART |
| Intracellular Domain | 1 | 2611-2785 | Phobius, SMART |
Expression
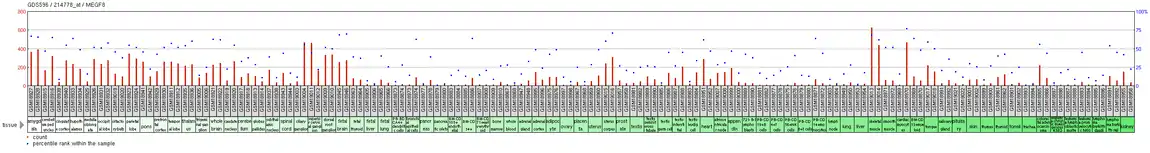
MEGF8 is found to be expressed at high levels in cardiac myocytes and fetal brain tissue, according to GeoProfiles,[23] from NCBI. This GeoProfile also indicated that MEGF8 was found to be at moderate to moderately low expression levels in all other tissues examined. NCBI GeoProfile data also provided the tissue expression graph for MEGF8 in humans, which is displayed to the right, further illustrating specific sites and levels of expression[24]
Molecular Function
According to BioGPS[25] gene ontology information, MEGF8 is an active participant in receptor activity, calcium ion binding, protein binding.
Role in Biological Processes
Analysis of gene ontology information by BioGPS[25] was able to produce a list of biological processes in each of which MEGF8 plays a significant role:
- embryonic heart tube morphogenesis (GO:0003143)
- regulation of gene expression (GO:0010468)
- embryonic limb morphogenesis (GO:0030326)
- BMP signaling pathway (GO:0030509)
- limb morphogenesis (GO:0035108)
- cell migration involved in gastrulation (GO:0042074)
- embryonic skeletal system morphogenesis (GO:0048704)
- positive regulation of axon extension involved in axon guidance (GO:0048842)
- epiboly involved in gastrulation with mouth forming second (GO:0055113)
- embryonic heart tube left/right pattern formation (GO:0060971)
- left/right pattern formation (GO:0060972)
- determination of heart left/right asymmetry (GO:0061371)
- determination of digestive tract left/right asymmetry (GO:0071907)
- craniofacial suture morphogenesis (GO:0097094)
- fasciculation of sensory neuron axon (GO:0097155)
Putative Interactions
In the table below, all predicted interactions, except SMARCD3, are supported by two-hybrid screen experimental data. This information is supported by both NextProt[26] database and IntAct database.[27] The two interactions with the highest confidence value are also supported by materials found by text-mining in STRING.[28] Together, it is with reasonably high confidence that the proteins in red are interacting with MEGF8, and with moderate confidence that the proteins in green interact with MEGF8. The confidence level for the proteins in blue is much lower, which may mean that the two-hybrid assay provided a false positive, or that they actually are interacting.
| Predicted Interacting Protein | Confidence | Location | Description | Experimental/Text Support | Function | Source | |
| GFI1B | Conf:0.866 | Found in Endothelial & Erythroid | GFI1B is a growth factor independent 1B transcription repressor | Two-Hybrid (IntAct) Text-mining (STRING/OMIM) | Essential proto-oncogenic transcriptional regulator; Transcriptional repressor or activator depending on both promoter and cell type context; represses promoter activity of SOCS1 and SOCS3 and thus, may regulate cytokine signaling pathways. | IntAct, STRING, NextProt | |
| ATN1 | Conf: 0.538 | Everywhere | Atrophin 1 (ATN1) | Two Hybrid Assay | Transcriptional corepressor. Recruits NR2E1 to repress transcription. Promotes vascular smooth cell (VSMC) migration and orientation | IntAct, STRING | |
| ATXN7 | Conf: 0.510 | Mod-High Everywhere | Apinocerebellar ataxia type 7 protein (ATXN7) | Two Hybrid, Pull-Down | Acts as component of the STAGA transcription coactivator-HAT complex. Mediates the interaction of STAGA complex with the CRX and is involved in CRX-dependent gene activation. Necessary for microtubule cytoskeleton stabilization | Int Act, NextProt | |
| CACNA1A | Conf: 0.510 | Certain Brain Tissues | Calcium Channel, Voltage-Dependent, P/Q Type, Alpha 1A Subunit (Cav2.1) | Two Hybrid Assay, Pull-Down | Mediates the entry of calcium ions into excitable cells and are also involved in a variety of calcium-dependent processes, including muscle contraction, hormone or neurotransmitter release, gene expression, cell motility, cell division and cell death. | IntAct, NextProt | |
| SMARCD3 | Conf: 0.778 | High Everywhere | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member (SMARCD3) | Text-mining (OMIM article for SMARCS3) | Plays a role in ATP dependent nucleosome remodeling by SMARCA4 containing complexes. Stimulates nuclear receptor mediated transcription | STRING | |
| FIHB1 | Conf: 0.370 | Two Hybrid Pooling | Uncharacterized | IntAct, NextProt | |||
| Y3542 | Conf: 0.370 | (Q8CKF8 in UniProtKB) | Two Hybrid Pooling | Uncharacterized | IntAct, NextProt | ||
| ProW | Conf: 0.370 | Two Hybrid Pooling | Uncharacterized | IntAct, NextProt |
Alternative Splicing, Mutations, & Phenotypic Impacts
Splice Variants
The four primary splice variants and their distinctions are described below (labels correspond to those in image below):
A: has spliced out Exon 13. Looking at the attached working conceptual translation, it can be seen that exon 3 does not code for any feature, domain, motif or other functional section of aa, and is likely therefore not key to the function of MEGF8 protein. This is the variant that corresponds to the splice model of the analyzed megf8.
B: Spliced out exons 1-6; these exons hold several key domains and motifs including the CUB domain, two PSI domains, a D1k3ia3 structural domain, and a kelch repeat. This may result in a misfolded protein without the structural segments, and inhibit participation in development events (loss of PSI and CUB). Still has signal and TMEM so may still be able to partially function
C: part of the D1k3ia3 structural domain remains in exon 29, but the kelch repeat has been excised, which could lead to structural issues. Also this variant contains almost 3 PSI domains, and an area of low complexity in exons 32-35, which may allow this variant to function in the cell, but no signal or TMEM to place in membrane so not a normal function
D: This variant is exons 36-40, excised 41, and a shortened 42 exon. It possesses EGF calcium domains and EGF/EGF-like domains. Loss of 41 will drastically alter the function as it possesses the TMEM segment. It depends on where 41 is lost and 42 is cleaved.

SNPs
There are several SNPs, found through NCBI GeneView,[29] that cause missense or silent mutations in MEGF8. However, three SNP mutations were identified as causes of Carpenter Syndrome 2 by Twigg et al.[30] The three SNP mutations are: Gly199 to Arg; Arg1499 to His; Ser2367 to Gly. The article by Twigg includes a supplementary data set that shows a multiple sequence alignment of the regions surrounding the SNPs and the domain in which the SNP lies. The Gly199 to Arg mutation is located inside an EGF-domain; the Arg1499 to His mutation is located within a kelch domain in the 7-bladed beta-sheet propeller; the Ser2367 to Gly is located within an EGF-Laminin domain. These domain are important to maintaining a properly folded protein and its function.
Carpenter Syndrome 2
Visit Carpenter syndrome for more extensive details related to the disease. Genetic mutations in MEGF8 have been found to be a principal cause of this rare genetic syndrome.
Adverse Phenotypic Consequences
Mutations in MEGF8 have been found to be linked to defective lateralization during development, as reported by Twigg et al.[30] Common features of individuals with Carpenter Syndrome Subtype II include the following:
- Tower-shaped skull (craniosynostosis)[31]
- Mental Retardation[31]
- Polysyndactyly Digits[31]
- High birth weight[32]
- Obesity in later life[32] *congenital heart disease
- Umbilical hernia[32]
- Cryptorchidism in males[32]
- Genu valgum ("knock-knee")[30]
Current Research
There is no research being done currently to develop treatment or cures for Carpenter Syndrome 2. Researchers are still striving to understand the cause of the point mutations in MEGF8 that result in this extremely rare genetic disease.
References
- GRCh38: Ensembl release 89: ENSG00000105429 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000045039 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Zhang Z, Alpert D, Francis R, Chatterjee B, Yu Q, Tansey T, Sabol SL, Cui C, Bai Y, Koriabine M, Yoshinaga Y, Cheng JF, Chen F, Martin J, Schackwitz W, Gunn TM, Kramer KL, De Jong PJ, Pennacchio LA, Lo CW (March 2009). "Massively parallel sequencing identifies the gene Megf8 with ENU-induced mutation causing heterotaxy". Proceedings of the National Academy of Sciences of the United States of America. 106 (9): 3219–24. Bibcode:2009PNAS..106.3219Z. doi:10.1073/pnas.0813400106. PMC 2651267. PMID 19218456.
- "Multiple Epidermal Growth Factor-like Domains 8 Gene". MEGF8 Gene Card. Weizmann Institute of Science in Israel. Retrieved 4 February 2014.
- Schultz J, Milpetz F, Bork P, Ponting CP (May 1998). "SMART, a simple modular architecture research tool: identification of signaling domains". Proceedings of the National Academy of Sciences of the United States of America. 95 (11): 5857–64. Bibcode:1998PNAS...95.5857S. doi:10.1073/pnas.95.11.5857. PMC 34487. PMID 9600884.
- "BLAST". Retrieved 8 March 2014.
- Brendel, Volker. "Statistical Analysis of PS". SAPS. Department of Mathematics, Stanford University. Retrieved 27 April 2014.
- Pappas Jr.., Georgios J. "PELE-Protein Structure Prediction". SDSC Biology Workbench. Retrieved 24 April 2014.
- Kelley LA, Sternberg MJ (26 February 2009). "Protein structure prediction on the web: a case study using the Phyre server". pp. 363–371. Retrieved 27 April 2014.
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (January 2011). "CDD: a Conserved Domain Database for the functional annotation of proteins". Nucleic Acids Research. 39 (Database issue): D225–9. doi:10.1093/nar/gkq1189. PMC 3013737. PMID 21109532.
- Davis CG (May 1990). "The many faces of epidermal growth factor repeats". The New Biologist. 2 (5): 410–9. PMID 2288911.
- Nakai K, Horton P (January 1999). "PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization". Trends in Biochemical Sciences. 24 (1): 34–6. doi:10.1016/S0968-0004(98)01336-X. PMID 10087920.
- Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG (2008). "Data growth and its impact on the SCOP database: new developments". Nucleic Acids Research. 36 (Database issue): D419–25. doi:10.1093/nar/gkm993. PMC 2238974. PMID 18000004.
- Ceroni A, Passerini A, Vullo A, Frasconi P (July 2006). "DISULFIND: a disulfide bonding state and cysteine connectivity prediction server". Nucleic Acids Research. 34 (Web Server issue): W177–81. doi:10.1093/nar/gkl266. PMC 1538823. PMID 16844986.
- Gramatikoff, Kosi; et al. (2004). "In Frontiers of Biotechnology and Pharmaceuticals". Science Press (4): 181.
- Blom N, Gammeltoft S, Brunak S (December 1999). "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites". Journal of Molecular Biology. 294 (5): 1351–62. doi:10.1006/jmbi.1999.3310. PMID 10600390.
- Karlin S (June 1995). "Statistical significance of sequence patterns in proteins". Current Opinion in Structural Biology. 5 (3): 360–71. doi:10.1016/0959-440X(95)80098-0. PMID 7583634.
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S (June 2004). "Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence". Proteomics. 4 (6): 1633–49. doi:10.1002/pmic.200300771. PMID 15174133. S2CID 18810164.
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (July 2004). "Improved prediction of signal peptides: SignalP 3.0". Journal of Molecular Biology. 340 (4): 783–95. CiteSeerX 10.1.1.165.2784. doi:10.1016/j.jmb.2004.05.028. PMID 15223320.
- Käll L, Krogh A, Sonnhammer EL (July 2007). "Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server". Nucleic Acids Research. 35 (Web Server issue): W429–32. doi:10.1093/nar/gkm256. PMC 1933244. PMID 17483518.
- "MEGF8 - Large-scale analysis of the human transcriptome (HG-U133A)". GeoProfiles. NCBI. Retrieved 15 April 2014.
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A (January 2011). "NCBI GEO: archive for functional genomics data sets--10 years on". Nucleic Acids Research. 39 (Database issue): D1005–10. doi:10.1093/nar/gkq1184. PMC 3013736. PMID 21097893.
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, Su AI (2009). "BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources". Genome Biology. 10 (11): R130. doi:10.1186/gb-2009-10-11-r130. PMC 3091323. PMID 19919682.
- Lane L, Argoud-Puy G, Britan A, Cusin I, Duek PD, Evalet O, Gateau A, Gaudet P, Gleizes A, Masselot A, Zwahlen C, Bairoch A (January 2012). "neXtProt: a knowledge platform for human proteins". Nucleic Acids Research. 40 (Database issue): D76–83. doi:10.1093/nar/gkr1179. PMC 3245017. PMID 22139911.
- Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H (January 2012). "The IntAct molecular interaction database in 2012". Nucleic Acids Research. 40 (Database issue): D841–6. doi:10.1093/nar/gkr1088. PMC 3245075. PMID 22121220.
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C (January 2009). "STRING 8--a global view on proteins and their functional interactions in 630 organisms". Nucleic Acids Research. 37 (Database issue): D412–6. doi:10.1093/nar/gkn760. PMC 2686466. PMID 18940858.
- Maglott D, Ostell J, Pruitt KD, Tatusova T (January 2011). "Entrez Gene: gene-centered information at NCBI". Nucleic Acids Research. 39 (Database issue): D52–7. doi:10.1093/nar/gkq1237. PMC 3013746. PMID 21115458.
- Twigg SR, Lloyd D, Jenkins D, Elçioglu NE, Cooper CD, Al-Sannaa N, Annagür A, Gillessen-Kaesbach G, Hüning I, Knight SJ, Goodship JA, Keavney BD, Beales PL, Gileadi O, McGowan SJ, Wilkie AO (November 2012). "Mutations in multidomain protein MEGF8 identify a Carpenter syndrome subtype associated with defective lateralization". American Journal of Human Genetics. 91 (5): 897–905. doi:10.1016/j.ajhg.2012.08.027. PMC 3487118. PMID 23063620.
- Frias JL, Felman AH, Rosenbloom AL, Finkelstein SN, Hoyt WF, Hall BD (1978). "Normal intelligence in two children with Carpenter syndrome". American Journal of Medical Genetics. 2 (2): 191–9. doi:10.1002/ajmg.1320020210. PMID 263437.
- Cohen DM, Green JG, Miller J, Gorlin RJ, Reed JA (October 1987). "Acrocephalopolysyndactyly type II--Carpenter syndrome: clinical spectrum and an attempt at unification with Goodman and Summit syndromes". American Journal of Medical Genetics. 28 (2): 311–24. doi:10.1002/ajmg.1320280208. PMID 3322002.
Further reading
- Victorine AS, Weida J, Hines KA, Robinson B, Torres-Martinez W, Weaver DD (March 2014). "Prenatal diagnosis of Carpenter syndrome: looking beyond craniosynostosis and polysyndactyly". American Journal of Medical Genetics. Part A. 164A (3): 820–3. doi:10.1002/ajmg.a.36362. PMID 24458945. S2CID 3040251.
- Engelhard C, Sarsfield S, Merte J, Wang Q, Li P, Beppu H, Kolodkin AL, Sucov HM, Ginty DD (September 2013). "MEGF8 is a modifier of BMP signaling in trigeminal sensory neurons". eLife. 2: e01160. doi:10.7554/eLife.01160. PMC 3776557. PMID 24052814.
- Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O (July 1998). "Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening". Genomics. 51 (1): 27–34. doi:10.1006/geno.1998.5341. PMID 9693030.
- Zhong J, Zou H (August 2014). "BMP signaling in axon regeneration". Current Opinion in Neurobiology. 27: 127–34. doi:10.1016/j.conb.2014.03.009. PMC 4122622. PMID 24713578.
- Twigg SR, Lloyd D, Jenkins D, Elçioglu NE, Cooper CD, Al-Sannaa N, Annagür A, Gillessen-Kaesbach G, Hüning I, Knight SJ, Goodship JA, Keavney BD, Beales PL, Gileadi O, McGowan SJ, Wilkie AO (November 2012). "Mutations in multidomain protein MEGF8 identify a Carpenter syndrome subtype associated with defective lateralization". American Journal of Human Genetics. 91 (5): 897–905. doi:10.1016/j.ajhg.2012.08.027. PMC 3487118. PMID 23063620.
- Zhang Z, Alpert D, Francis R, Chatterjee B, Yu Q, Tansey T, Sabol SL, Cui C, Bai Y, Koriabine M, Yoshinaga Y, Cheng JF, Chen F, Martin J, Schackwitz W, Gunn TM, Kramer KL, De Jong PJ, Pennacchio LA, Lo CW (March 2009). "Massively parallel sequencing identifies the gene Megf8 with ENU-induced mutation causing heterotaxy". Proceedings of the National Academy of Sciences of the United States of America. 106 (9): 3219–24. Bibcode:2009PNAS..106.3219Z. doi:10.1073/pnas.0813400106. PMC 2651267. PMID 19218456.