Oligosaccharyltransferase
Oligosaccharyltransferase or OST (EC 2.4.1.119) is a membrane protein complex that transfers a 14-sugar oligosaccharide from dolichol to nascent protein. It is a type of glycosyltransferase. The sugar Glc3Man9GlcNAc2 (where Glc=Glucose, Man=Mannose, and GlcNAc=N-acetylglucosamine) is attached to an asparagine (Asn) residue in the sequence Asn-X-Ser or Asn-X-Thr where X is any amino acid except proline. This sequence is called a glycosylation sequon. The reaction catalyzed by OST is the central step in the N-linked glycosylation pathway.
| STT3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
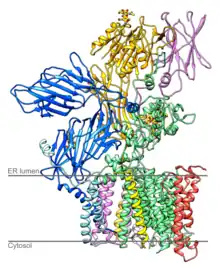 Structure of yeast OST [1] | |||||||||
| Identifiers | |||||||||
| Symbol | STT3 | ||||||||
| Pfam | PF02516 | ||||||||
| Pfam clan | CL0111 | ||||||||
| InterPro | IPR003674 | ||||||||
| OPM superfamily | 242 | ||||||||
| OPM protein | 3rce | ||||||||
| CAZy | GT66 | ||||||||
| Membranome | 275 | ||||||||
| |||||||||
Location
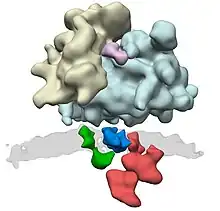
OST is a component of the translocon in the endoplasmic reticulum (ER) membrane. A lipid-linked core-oligosaccharide is assembled at the membrane of the endoplasmic reticulum and transferred to selected asparagine residues of nascent polypeptide chains by the oligosaccharyl transferase complex.[3] The active site of OST is located about 4 nm from the lumenal face of the ER membrane.[4]
It usually acts during translation as the nascent protein is entering the ER, but this cotranslational glycosylation is nevertheless called a posttranslational modification. A few examples have been found of OST activity after translation is complete.[5][6] Current opinion is that post-translational activity may occur if the protein is poorly folded or folds slowly.[6]
Structure and function
Yeast OST is composed of eight different membrane-spanning proteins in three subcomplexes (one of them is OST4).[7][8] These octomers do not form higher order oligomers, and three of the eight proteins are glycosylated themselves.[7] OST in mammals is known to have a similar composition.[9][10]
OST is thought to require many subunits because it must:[11]
- Position itself near the translocon pore.
- Recognize and bind oligosaccharyldolichol.
- Scan the nascent protein in order to recognize and bind sequons.
- Move these two large substrates into their proper locations and conformations.
- Activate the Asn amide nitrogen atom for the actual transfer of oligosaccharide.
- Release its substrates.
The catalytically active subunit of the OST is called STT3. Two paralogs exist in eukaryotes, termed STT3A and STT3B. STT3A is primarily responsible for cotranslational glycosylation of the nascent polypeptide as it enters the lumen of the endoplasmic reticulum whereas STT3B can also mediate posttranslational glycosylation.[12] The structure of PglB, the prokaryotic homolog of STT3 has been solved.[13] The high sequence similarity between the prokaryotic and the eukaryotic STT3 suggests that their structures are similar.
Clinical significance
CDG syndromes are genetic disorders of the glycosylation pathway. They are labelled "Type I" if the defective gene is for an enzyme involved in the assembly or transfer of the Glc3Man9GlcNAc2-dolichol precursor. They are labelled "Type II" if the defective step occurs after the action of OST in the N-linked glycosylation pathway or involves O-linked glycosylation.[14]
References
- Wild R, Kowal J, Eyring J, Ngwa EM, Aebi M, Locher KP (2018). "Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation". Science. 359 (6375): 545–550. Bibcode:2018Sci...359..545W. doi:10.1126/science.aar5140. PMID 29301962.
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Förster F (2014). "Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon". Nat. Commun. 5 (5): 3072. Bibcode:2014NatCo...5.3072P. doi:10.1038/ncomms4072. PMID 24407213.
- Zufferey R, Knauer R, Burda P, Stagljar I, te Heesen S, Lehle L, Aebi M (October 1995). "STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo". EMBO J. 14 (20): 4949–60. doi:10.1002/j.1460-2075.1995.tb00178.x. PMC 394598. PMID 7588624.
- Nilsson IM, von Heijne G (March 1993). "Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane". J. Biol. Chem. 268 (8): 5798–801. doi:10.1016/S0021-9258(18)53389-5. PMID 8449946.
- Pless DD, Lennarz WJ; Lennarz (January 1977). "Enzymatic conversion of proteins to glycoproteins". Proc. Natl. Acad. Sci. U.S.A. 74 (1): 134–8. Bibcode:1977PNAS...74..134P. doi:10.1073/pnas.74.1.134. PMC 393212. PMID 264667.
- Duvet S, Op De Beeck A, Cocquerel L, Wychowski C, Cacan R, Dubuisson J (February 2002). "Glycosylation of the hepatitis C virus envelope protein E1 occurs posttranslationally in a mannosylphosphoryldolichol-deficient CHO mutant cell line". Glycobiology. 12 (2): 95–101. doi:10.1093/glycob/12.2.95. PMID 11886842.
- Knauer R, Lehle L (January 1999). "The oligosaccharyltransferase complex from yeast". Biochim. Biophys. Acta. 1426 (2): 259–73. doi:10.1016/S0304-4165(98)00128-7. PMID 9878773.
- Dempski RE, Imperiali B (December 2002). "Oligosaccharyl transferase: gatekeeper to the secretory pathway". Curr Opin Chem Biol. 6 (6): 844–50. doi:10.1016/S1367-5931(02)00390-3. PMID 12470740.
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, von Heijne G, Johnson AE (May 2003). "Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex". J. Cell Biol. 161 (4): 715–25. doi:10.1083/jcb.200301043. PMC 2199356. PMID 12756234.
- Karaoglu D, Kelleher DJ, Gilmore R (October 2001). "Allosteric regulation provides a molecular mechanism for preferential utilization of the fully assembled dolichol-linked oligosaccharide by the yeast oligosaccharyltransferase". Biochemistry. 40 (40): 12193–206. doi:10.1021/bi0111911. PMID 11580295.
- Imperiali B (November 1997). "Protein Glycosylation: The Clash of the Titans". Accounts of Chemical Research. 30 (11): 452–459. doi:10.1021/ar950226k.
- Ruiz-Canada C, Kelleher DJ, Gilmore R (January 2009). "Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms". Cell. 136 (2): 272–83. doi:10.1016/j.cell.2008.11.047. PMC 2859625. PMID 19167329.
- Lizak C, Gerber S, Numao S, Aebi M, Locher KP (June 2011). "X-ray structure of a bacterial oligosaccharyltransferase". Nature. 474 (7351): 350–5. doi:10.1038/nature10151. PMID 21677752. S2CID 205225231.
- Marquardt T, Denecke J (June 2003). "Congenital disorders of glycosylation: review of their molecular bases, clinical presentations and specific therapies". Eur. J. Pediatr. 162 (6): 359–79. doi:10.1007/s00431-002-1136-0. PMID 12756558. S2CID 3196550.
External links
- oligosaccharyltransferase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)