Ribonuclease T
Ribonuclease T (RNase T, exonuclease T, exo T) is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria,[2] as well as in DNA repair pathways.[3] It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.[1][2]
| Ribonuclease T | |||||||||
|---|---|---|---|---|---|---|---|---|---|
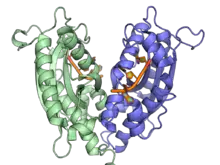 A ribonuclease T dimer in complex with DNA (orange), from PDB ID 3NH1.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | rnt | ||||||||
| Pfam | PF00929 | ||||||||
| InterPro | IPR013520 | ||||||||
| SMART | SM00479 | ||||||||
| |||||||||
Structure and mechanism
RNAse T catalyzes the removal of nucleotides from the 3' end of both RNA and DNA. It is inhibited by both double stranded DNA and RNA, as well as cytosine residues on the 3' end of RNA. Two cytosines at the 3' end of RNA appear to remove the activity of RNAse T entirely.[3] This cytosine effect, however, is observed less with ssDNA. This lack of sequence specificity in ssDNA, combined with its ability to act on ssDNA close to a duplex region, has led to its use in creating blunt ends for DNA cloning.[4] Structurally, RNAse T exists as an anti-parallel dimer[5][6] and requires a divalent cation to function.[7]
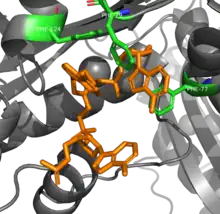
RNAse T is able to achieve its sequence specificity in RNA digestion via several aromatic residues that sandwich between nucleobases. The π-π interactions between four phenylalanine residues and the two nucleotides at the 3' end are different depending on the identify of the nucleotides, which changes the conformation and thus activity of the enzyme.[8] An additional glutamic acid residue rotates to hydrogen bond to cytosine by not other bases, further increasing specificity.[9]
Function
A member of the larger DEDD family of exoribonucleases, RNAse T plays a key role in the maturation of tRNA[10] as well as the maturation of the 5S[11] and 23S[12] rRNA domains. Specifically, RNAse T cleaves the 3' AMP residue from the 3' CCA sequences at the end of tRNA, which explains RNAse T's sequence specificity for stopping at the 3' CC sequence.[13] Additionally, RNAse T can play a role in DNA repair by cleaving the 3' end of bulge DNA.[3]
While E. coli can survive without RNAse T, its absence leads to slower life cycles and weakened response to starvation.[14] Additionally, the presence of RNAse T in E. coli is linked to increased resistance to UV damage.[15] It has been theorized that, while other ribonculeases can perform the function RNAse T, the fact that RNAse T is more effective at cleaving DNA and RNA near double-stranded regions means that alternatives are less effective.[16] Despite the apparent usefulness of RNAse T, the enzyme is only found in gammaproteobacteria.[17]
In E. coli, RNAse T is encoded by the rnt gene and is hypothesized to have diverged from the proofreading subunits of polymerase III during the emergence of gammaproteobacteria.[16][17]
References
- Hsiao YY, Duh Y, Chen YP, Wang YT, Yuan HS (September 2012). "How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes". Nucleic Acids Research. 40 (16): 8144–54. doi:10.1093/nar/gks548. PMC 3439924. PMID 22718982.
- Zuo Y, Deutscher MP (August 2002). "The physiological role of RNase T can be explained by its unusual substrate specificity". The Journal of Biological Chemistry. 277 (33): 29654–61. doi:10.1074/jbc.M204252200. PMID 12050169.
- Hsiao YY, Fang WH, Lee CC, Chen YP, Yuan HS (March 2014). "Structural insights into DNA repair by RNase T--an exonuclease processing 3' end of structured DNA in repair pathways". PLOS Biology. 12 (3): e1001803. doi:10.1371/journal.pbio.1001803. PMC 3942315. PMID 24594808.
- Zuo Y, Deutscher MP (October 1999). "The DNase activity of RNase T and its application to DNA cloning". Nucleic Acids Research. 27 (20): 4077–82. doi:10.1093/nar/27.20.4077. PMC 148676. PMID 10497273.
- Li Z, Zhan L, Deutscher MP (January 1996). "Escherichia coli RNase T functions in vivo as a dimer dependent on cysteine 168". The Journal of Biological Chemistry. 271 (2): 1133–7. doi:10.1074/jbc.271.2.1133. PMID 8557641.
- Zuo Y, Zheng H, Wang Y, Chruszcz M, Cymborowski M, Skarina T, et al. (April 2007). "Crystal structure of RNase T, an exoribonuclease involved in tRNA maturation and end turnover". Structure. 15 (4): 417–28. doi:10.1016/j.str.2007.02.004. PMC 1907377. PMID 17437714.
- Deutscher MP, Marlor CW (June 1985). "Purification and characterization of Escherichia coli RNase T". The Journal of Biological Chemistry. 260 (11): 7067–71. doi:10.1016/S0021-9258(18)88888-3. PMID 3888994.
- Duh Y, Hsiao YY, Li CL, Huang JC, Yuan HS (December 2015). "Aromatic residues in RNase T stack with nucleobases to guide the sequence-specific recognition and cleavage of nucleic acids". Protein Science. 24 (12): 1934–41. doi:10.1002/pro.2800. PMC 4815224. PMID 26362012.
- Hsiao YY, Yang CC, Lin CL, Lin JL, Duh Y, Yuan HS (April 2011). "Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation". Nature Chemical Biology. 7 (4): 236–43. doi:10.1038/nchembio.524. PMID 21317904.
- Li Z, Deutscher MP (August 1996). "Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo". Cell. 86 (3): 503–12. doi:10.1016/s0092-8674(00)80123-3. PMID 8756732.
- Li Z, Deutscher MP (July 1995). "The tRNA processing enzyme RNase T is essential for maturation of 5S RNA". Proceedings of the National Academy of Sciences of the United States of America. 92 (15): 6883–6. Bibcode:1995PNAS...92.6883L. doi:10.1073/pnas.92.15.6883. PMC 41434. PMID 7542780.
- Li Z, Pandit S, Deutscher MP (January 1999). "Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T". RNA. 5 (1): 139–46. doi:10.1017/s1355838299981669. PMC 1369746. PMID 9917073.
- Deutscher MP, Marlor CW, Zaniewski R (July 1984). "Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA". Proceedings of the National Academy of Sciences of the United States of America. 81 (14): 4290–3. Bibcode:1984PNAS...81.4290D. doi:10.1073/pnas.81.14.4290. PMC 345573. PMID 6379642.
- Kelly KO, Deutscher MP (October 1992). "The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli". Journal of Bacteriology. 174 (20): 6682–4. doi:10.1128/jb.174.20.6682-6684.1992. PMC 207653. PMID 1400219.
- Viswanathan M, Lanjuin A, Lovett ST (March 1999). "Identification of RNase T as a high-copy suppressor of the UV sensitivity associated with single-strand DNA exonuclease deficiency in Escherichia coli". Genetics. 151 (3): 929–34. doi:10.1093/genetics/151.3.929. PMC 1460521. PMID 10049912.
- Bechhofer DH, Deutscher MP (June 2019). "Bacterial ribonucleases and their roles in RNA metabolism". Critical Reviews in Biochemistry and Molecular Biology. 54 (3): 242–300. doi:10.1080/10409238.2019.1651816. PMC 6776250. PMID 31464530.
- Zuo Y, Deutscher MP (March 2001). "Exoribonuclease superfamilies: structural analysis and phylogenetic distribution". Nucleic Acids Research. 29 (5): 1017–26. doi:10.1093/nar/29.5.1017. PMC 56904. PMID 11222749.