STK3
Serine/threonine-protein kinase 3 is an enzyme that in humans is encoded by the STK3 gene.[5][6]
Background
Protein kinase activation is a frequent response of cells to treatment with growth factors, chemicals, heat shock, or apoptosis-inducing agents. This protein kinase activation presumably allows cells to resist unfavorable environmental conditions. The yeast 'sterile 20' (Ste20) kinase acts upstream of the mitogen-activated protein kinase (MAPK) cascade that is activated under a variety of stress conditions. MST2 was first identified as a kinase that resembles budding yeast Ste20 (Creasy and Chernoff, 1996) and later as a kinase that is activated by the proapoptotic agents straurosporine and FAS ligand (MIM 134638) (Taylor et al., 1996; Lee et al., 2001).[supplied by OMIM][6]
Structure
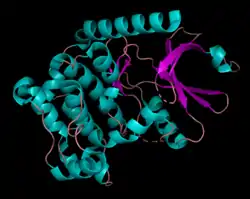
Human serine/threonine-protein kinase 3 (STK3, or MST2) is a 56,301 Da[7] monomer with three domains: a SARAH domain, composed of a long α-helix at the C-terminus that when dimerized, forms an antiparallel dimeric coiled-coil, an inhibitory domain, and a catalytic kinase domain at the N-terminus.[8] The SARAH (Salvador/RASSF/Hpo) domain has been found to mediate dimeric interactions between MST2 and RASSF enzymes, a class of tumor suppressors that serve an important role in activating apoptosis, as well as between MST2 and SAV1, a non-catalytic polypeptide responsible for bringing MST2 to an apoptotic pathway.[9][10] When the MST2 kinase domain is in its active state, a threonine residue residing on an alpha helix at the 180th position (T180) is autophosphorylated.[11]

Mechanism
Activation
STK3 is activated through autophosphorylation by dimerizing with itself or heterodimerizing with its homolog, MST1 (STK4).[12] Heterodimerization has been shown to exhibit a roughly six-fold weaker binding affinity than homodimerization with MST2, as well as lower kinase activity compared to both MST2/MST2 and MST1/MST1 homodimers.[10] In addition to activation by straurosporine and FAS ligand, STK3 has been found to be activated through dissociation of GLRX and Thioredoxin (Trx1) from STK3 under oxidative stress.[12] Recent studies have shown that when caspase 3 is activated during apoptosis, MST2 is cleaved, resulting in removal of the regulatory SARAH and inhibitory domains and thus regulation of MST2's kinase activity. Because cleavage by caspase 3 also cleaves off MST2's nuclear export signal, the MST2 kinase fragment can diffuse into the nucleus and phosphorylate Ser14 of histone H2B, promoting apoptosis.[10]
Inactivation
Inactivation of MST2 can be accomplished in several ways, including inhibition of MST2 homodimerization and autophosphorylation by c-Raf, which binds to the MST2 SARAH domain,[10] and phosphorylation of the highly conserved Thr117 by Akt (protein kinase B), blocking autophosphorylation of Thr180, MST2 cleavage, kinase activity, and translocation to the nucleus.[13]
MST2 substrates
In the mammalian Hippo signaling pathway, MST2, along with its homolog MST1, serves as an upstream kinase whose catalytic activity is responsible for downstream events leading to downregulation of proliferation-associated genes and increased transcription of proapoptotic genes.[12] When MST2 binds to SAV1 through its SARAH domain, MST2 phosphorylates LATS1/LATS2 with the help of SAV1, MOB1A/MOB1B, and Merlin (protein). In turn, LATS1/LATS2 phosphorylates and inhibits YAP1, preventing its movement into the nucleus and activation of transcription of pro-proliferative, anti-apoptotic and migration-associated genes. In the cytoplasm, YAP1 is marked for degradation by the SCF complex.[14] Additionally, MST2 phosphorylates transcription factors in the FOXO (Forkhead box O) family, which diffuse into the nucleus and activate transcription of pro-apoptotic genes.[12]
Disease Relevance
In many types of cancers, the proto-oncogene c-Raf binds to the SARAH domain of MST2 and prevents RASSF1A-mediated MST2 dimerization and subsequent downstream pro-apoptotic signaling.[15] Research has shown that in cells with loss of PTEN (gene), a tumor suppressor that is frequently mutated in cancers, Akt activity is upregulated, resulting in increased MST2 inactivation and undesirable cell proliferation.[16]
References
- GRCh38: Ensembl release 89: ENSG00000104375 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000022329 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Taylor LK, Wang HC, Erikson RL (September 1996). "Newly identified stress-responsive protein kinases, Krs-1 and Krs-2". Proceedings of the National Academy of Sciences of the United States of America. 93 (19): 10099–104. doi:10.1073/pnas.93.19.10099. PMC 38343. PMID 8816758.
- "Entrez Gene: STK3 serine/threonine kinase 3 (STE20 homolog, yeast)".
- "PhosphoSitePlus: Serine/threonine-protein kinase 3 - Protein Information".
- Liu G, Shi Z, Jiao S, Zhang Z, Wang W, Chen C, Hao Q, Hao Q, Zhang M, Feng M, Xu L, Zhang Z, Zhou Z, Zhang M (March 2014). "Structure of MST2 SARAH domain provides insights into its interaction with RAPL". Journal of Structural Biology. 185 (3): 366–74. doi:10.1016/j.jsb.2014.01.008. PMID 24468289.
- Sánchez-Sanz G, Tywoniuk B, Matallanas D, Romano D, Nguyen LK, Kholodenko BN, Rosta E, Kolch W, Buchete NV (October 2016). "SARAH Domain-Mediated MST2-RASSF Dimeric Interactions". PLOS Computational Biology. 12 (10): e1005051. doi:10.1371/journal.pcbi.1005051. PMC 5055338. PMID 27716844.
- Galan JA, Avruch J (Sep 2016). "MST1/MST2 Protein Kinases: Regulation and Physiologic Roles". Biochemistry. 55 (39): 5507–5519. doi:10.1021/acs.biochem.6b00763. PMC 5479320. PMID 27618557.
- Ni L, et al. (Oct 2013). "Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5". Structure. 21 (10): 1757–1768. doi:10.1016/j.str.2013.07.008. PMC 3797246. PMID 23972470.
- Lessard-Beaudoin M, Laroche M, Loudghi A, Demers MJ, Denault JB, Grenier G, Riechers SP, Wanker EE, Graham RK (November 2016). "Organ-specific alteration in caspase expression and STK3 proteolysis during the aging process" (PDF). Neurobiology of Aging. 47: 50–62. doi:10.1016/j.neurobiolaging.2016.07.003. PMID 27552481. S2CID 3930860.
- Kim D, Shu S, Coppola MD, Kaneko S, Yuan Z, Cheng JQ (Mar 2010). "Regulation of Proapoptotic Mammalian ste20–Like Kinase MST2 by the IGF1-Akt Pathway". PLOS ONE. 5 (3): e9616. doi:10.1371/journal.pone.0009616. PMC 2834758. PMID 20231902.
- Meng Z, Moroishi T, Guan K (Jan 2016). "Mechanisms of Hippo pathway regulation". Genes Dev. 30 (1): 1–17. doi:10.1101/gad.274027.115. PMC 4701972. PMID 26728553.
- Nguyen LK, Matallanas DG, Romano D, Kholodenko BN, Kolch W (Jan 2015). "Competing to coordinate cell fate decisions: the MST2-Raf-1 signaling device". Cell Cycle. 14 (2): 189–199. doi:10.4161/15384101.2014.973743. PMC 4353221. PMID 25607644.
- Romano D, Matallanas D, Weitsman G, Preisinger C, Ng T, Kolch W (Feb 2010). "Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt". Cancer Res. 70 (3): 1195–1203. doi:10.1158/0008-5472.CAN-09-3147. PMC 2880716. PMID 20086174.
Further reading
- Creasy CL, Chernoff J (September 1995). "Cloning and characterization of a human protein kinase with homology to Ste20". The Journal of Biological Chemistry. 270 (37): 21695–700. doi:10.1074/jbc.270.37.21695. PMID 7665586.
- Schultz SJ, Nigg EA (October 1993). "Identification of 21 novel human protein kinases, including 3 members of a family related to the cell cycle regulator nimA of Aspergillus nidulans". Cell Growth & Differentiation. 4 (10): 821–30. PMID 8274451.
- Creasy CL, Chernoff J (December 1995). "Cloning and characterization of a member of the MST subfamily of Ste20-like kinases". Gene. 167 (1–2): 303–6. doi:10.1016/0378-1119(95)00653-2. PMID 8566796.
- Bren A, Welch M, Blat Y, Eisenbach M (September 1996). "Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY". Proceedings of the National Academy of Sciences of the United States of America. 93 (19): 10090–3. doi:10.1073/pnas.93.19.10090. PMC 38341. PMID 8816756.
- Wang HC, Fecteau KA (August 2000). "Detection of a novel quiescence-dependent protein kinase". The Journal of Biological Chemistry. 275 (33): 25850–7. doi:10.1074/jbc.M000818200. PMID 10840030.
- Lee KK, Ohyama T, Yajima N, Tsubuki S, Yonehara S (June 2001). "MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation". The Journal of Biological Chemistry. 276 (22): 19276–85. doi:10.1074/jbc.M005109200. PMID 11278283.
- De Souza PM, Kankaanranta H, Michael A, Barnes PJ, Giembycz MA, Lindsay MA (May 2002). "Caspase-catalyzed cleavage and activation of Mst1 correlates with eosinophil but not neutrophil apoptosis". Blood. 99 (9): 3432–8. doi:10.1182/blood.V99.9.3432. PMID 11964314. S2CID 8728566.
- Deng Y, Pang A, Wang JH (April 2003). "Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis". The Journal of Biological Chemistry. 278 (14): 11760–7. doi:10.1074/jbc.M211085200. PMID 12554736.
- Rabizadeh S, Xavier RJ, Ishiguro K, Bernabeortiz J, Lopez-Ilasaca M, Khokhlatchev A, Mollahan P, Pfeifer GP, Avruch J, Seed B (July 2004). "The scaffold protein CNK1 interacts with the tumor suppressor RASSF1A and augments RASSF1A-induced cell death". The Journal of Biological Chemistry. 279 (28): 29247–54. doi:10.1074/jbc.M401699200. PMID 15075335.
- O'Neill E, Rushworth L, Baccarini M, Kolch W (December 2004). "Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1". Science. 306 (5705): 2267–70. doi:10.1126/science.1103233. PMID 15618521. S2CID 30879956.
- Chan EH, Nousiainen M, Chalamalasetty RB, Schäfer A, Nigg EA, Silljé HH (March 2005). "The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1". Oncogene. 24 (12): 2076–86. doi:10.1038/sj.onc.1208445. PMID 15688006. S2CID 27285160.
- Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, Im CR, Lee JO, Yonehara S, Lim DS (March 2006). "Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis". Cancer Research. 66 (5): 2562–9. doi:10.1158/0008-5472.CAN-05-2951. PMID 16510573.
- Callus BA, Verhagen AM, Vaux DL (September 2006). "Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation". The FEBS Journal. 273 (18): 4264–76. doi:10.1111/j.1742-4658.2006.05427.x. PMID 16930133. S2CID 8261982.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (November 2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983. S2CID 7827573.
- Seidel C, Schagdarsurengin U, Blümke K, Würl P, Pfeifer GP, Hauptmann S, Taubert H, Dammann R (October 2007). "Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma". Molecular Carcinogenesis. 46 (10): 865–71. doi:10.1002/mc.20317. PMID 17538946. S2CID 36848574.
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'neill E (September 2007). "RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein". Molecular Cell. 27 (6): 962–75. doi:10.1016/j.molcel.2007.08.008. PMC 2821687. PMID 17889669.