Immunoglobulin M
Immunoglobulin M (IgM) is one of several isotypes of antibody (also known as immunoglobulin) that are produced by vertebrates. IgM is the largest antibody, and it is the first antibody to appear in the response to initial exposure to an antigen.[1][2] In humans and other mammals that have been studied, plasmablasts residing in the spleen are the main source for specific IgM production.[3][4]
| Immunoglobulin M | |||||||
|---|---|---|---|---|---|---|---|
| (pentamer) | |||||||
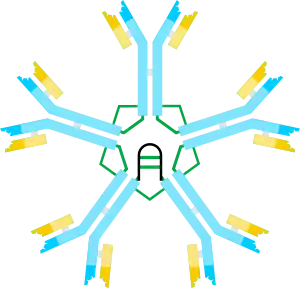 | |||||||
| Protein type | antibody | ||||||
| |||||||
History
In 1937, an antibody was observed in horses hyper-immunized with pneumococcus polysaccharide that was much larger than the typical rabbit γ-globulin,[5] with a molecular weight of 990,000 daltons.[6] In accordance with its large size, the new antibody was originally referred to as γ-macroglobulin, and then in subsequent terminology as IgM—M for “macro”. The V domains of normal immunoglobulin are highly heterogeneous, reflecting their role in protecting against the great variety of infectious microbes, and this heterogeneity impeded detailed structural analysis of IgM. Two sources of homogeneous IgM were subsequently discovered. First, the high molecular weight protein produced by some multiple myeloma patients was recognized to be a tumor-produced γ-macroglobulin, and because the tumor is a clone the IgM it produces is homogeneous.[7] In the 1960s, methods were developed for inducing immunoglobulin-producing tumors (plasmacytomas) in mice, thus providing a source of homogeneous immunoglobulins of various isotypes, including IgM (reviewed in[8]). More recently, expression of engineered immunoglobulin genes in tissue culture can be used to produce IgM with specific alterations and thus to identify the molecular requirements for features of interest.
Structure
Immunoglobulins are composed of light chains and heavy chains. The light chain (λ or κ) is a protein of ~220 amino acids, composed of a variable domain, VL (a segment of approximately 110 amino acids), and a constant domain, CL (also approximately 110 amino acids long). The µ heavy chain of IgM is a protein of ~576 amino acids, and includes a variable domain (VH ~110 amino acids), four distinct constant region domains (Cµ1, Cµ2, Cµ3, Cµ4, each ~110 amino acids) and a “tailpiece” of ~20 amino acids. The µ heavy chain bears oligosaccharides at five asparagine residues. The oligosaccharides on mouse and human IgM have been partially characterized by a variety of techniques, including NMR, lectin binding, various chromatographic systems and enzymatic sensitivity (reviewed in[9]). The structure of the oligosaccharides at each site varies in detail, and the predominant oligosaccharides – biantennary, triantennary, high mannose—differ among the sites.
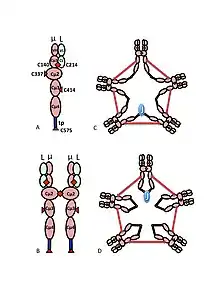
A) The µL heterodimer, sometimes called a halfmer, with variable (VH, VL) and constant region (Cµ1, Cµ2, Cµ3, Cµ4tp; CL) domains. The cysteines that mediate disulfide bonds between µ chains are shown as red arrowheads, so that a cysteine disulfide bond appears as a red double arrowhead (red diamond).
B) The IgM “monomer” (µL)2. The disulfide bonds between Cµ2 domains are represented by a red double arrowhead.
C, D) Two models for J chain-containing IgM pentamer that have appeared in various publications at various times. As in (B), the disulfide bonds between Cµ2 domains and the disulfide bonds between Cµ4tp domains are represented by a red double arrowhead; the Cµ3 disulfide bonds are represented (for clarity) by long double-headed arrows. The connectivity, i.e., the inter-chain disulfide bonding of the µ chains, is denoted like electrical connectivity. In (C) the Cµ3 disulfide bonds join µ chains in parallel with the Cµ4tp disulfide bonds, and these disulfide bonds join µ chains in series with the Cµ2 disulfide bonds. In (D) the Cµ2 and Cµ4tp disulfide bonds join µ chains in parallel and both types join µ chains in series with the Cµ3 disulfide bonds. (Figure reproduced with permission of the publisher and authors[10]).
The multimeric structure of IgM is shown schematically in Figure 1. Figure 1A shows the “heterodimer” composed of one light chain, denoted L, and one heavy chain, denoted µ. The heavy and light chains are held together both by disulfide bonds (depicted as red triangles) and by non-covalent interactions.
Figure 1B shows two µL units linked by a disulfide bond in the Cµ2 domains; this (µL)2 structure is often referred to as the IgM “monomer”, as it is analogous in some ways to the structure of immunoglobulin G (IgG).
On the basis of its sedimentation velocity and appearance in electron micrographs, it was inferred that IgM is mostly a “pentamer”, i.e., a polymer composed of five “monomers” [(µL)2]5, and was originally depicted by the models in Figures 1C and 1D, with disulfide bonds between the Cµ3 domains and between the tail pieces.[11][12] Also shown is that pentameric IgM includes a third protein, the J chain. J chain (J for joining) was discovered as a covalently bonded component of polymeric IgA and IgM.[13][14] J chain is a small (~137 amino acids), acidic protein. As shown, J chain joins two µ chains via disulfide bonds involving cysteines in the tailpieces.[15]
Molecular requirements for forming polymeric IgM
It was initially expected that the J chain would be important for forming the polymeric immunoglobulins, and indeed polymerization of IgA depends strongly (but not absolutely) on the J chain.[16][17] In contrast, polymeric IgM forms efficiently in the absence of the J chain.[18][19]
The predominant form of human and mouse IgM is the pentamer. By way of comparison, the structure of IgM from frog (Xenopus) is predominantly hexameric,[20][21] IgM from bony fish is predominantly tetrameric, and IgM from cartilaginous fish (shark) is predominantly pentameric.[22][23] Although the pentameric form predominates in mice and humans, the hexameric form has also been observed.[24][25] Subsequent studies using recombinant DNA expression systems indicated that the hexamer is a major form of mouse IgM when the IgM is produced under conditions where the incorporation of the J chain is prevented, either by producing IgM in cells that lack J chain[18] or by producing IgM with a µ heavy chain that lacks the cysteine in the tailpiece.[26][27] In summary, hexameric IgM never contains J chain; pentameric IgM can be formed so as to include or not include the J chain.[28]
An important difference between the µ and γ heavy chains is the availability of cysteines for forming disulfide bonds between heavy chains. In the case of the γ heavy chain, the only inter-γ bonds are formed by cysteines in the hinge, and accordingly, each γ chain binds to only one other γ chain. By contrast, the Cµ2 and Cµ3 domains and the tailpiece each include a cysteine that form a disulfide bond with another µ chain. The cysteines in the Cµ2 domains mediate the formation of monomeric IgM (µL)2. The tailpiece along with the included cysteine is necessary and sufficient for the formation of polymeric immunoglobulins. That is, deleting the tailpiece from the µ heavy chain prevents the formation of polymeric IgM.[29] Conversely, cells expressing a γ heavy chain that has been modified to include the tailpiece produce polymeric IgG.[30][31][32]
The role of the cysteine in the Cµ3 domain is more subtle. Figures 1C and 1D represent possible models for pentameric IgM. In these models, each µ chain is envisaged to bind two other µ chains. However, neither model alone can fully account for the structure of polymeric IgM. For example, the model in Figure 1C predicts that the disulfide bond between the Cµ2 domains is essential for making disulfide-bonded polymeric IgM. The model in Figure 1D predicts that the disulfide bond between the Cµ3 domains is essential. Disulfide bonded, polymeric, IgM can still be made if any one of the three cysteines is absent. In the context of models in which each µ chain interacts with only two other µ chains, these results suggest that some molecules are like Figure 1C and some like Figure 1D. However, the availability of three cysteines for inter-µ chain bonding suggests that the µ chains might each bind three other µ chains, as illustrated in Figure 2. In the same spirit, Figure 2C presents a model for J chain-containing pentamer that reflects evidence that J chain joins µ chains that are not joined to other µ chains by the cysteines in the Cµ3 domains. These and other models, both regular and irregular are discussed elsewhere.[27][33]
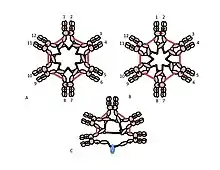
A, B) These figures depict two of many possible models of inter-µ chain disulfide bonding in hexameric IgM. As in Figure 1, the Cµ2 disulfide bonds and the Cµ4tp disulfide bonds are represented by a red double arrowhead, and the Cµ3 disulfide bonds are represented by the long double-headed arrows. In both models A and B each type of disulfide bond (Cµ2-Cµ2; Cµ3-Cµ3; Cµ4tp-Cµ4tp) joins µ chains in series with each of the others. Methods for distinguishing these and other models are discussed in reference [28].
C) This representation of pentameric IgM illustrates how J chain might be bonded to µ chains that are not linked via Cµ3 disulfide bonds
Pentameric IgM is typically represented as containing a single J chain per polymer, but in actuality the measurements of J chain stoichiometry have ranged from one J molecule per polymer to three J molecules per polymer.[34][35][36][37] The wide range might be due to technical problems, such as incomplete radiolabeling or imprecisely quantitating an Ouchterlony line. However, the variation might also be due to heterogeneity in the IgM preparations, i.e., the various preparations might have differed substantially in their content of J-containing and J-deficient polymers.
Tertiary and quaternary structure of the µ constant region
To gain insight into the detailed three-dimensional structure of the µ chain, the individual Cµ2, Cµ3, and Cµ4tp domains were produced separately in E. coli and then analyzed by a variety of methods, including sedimentation rate, X-ray crystallography, and NMR spectroscopy. As in the case of other immunoglobulins, the domains of the µ heavy chain have the characteristic overlying β-sheets comprising seven strands, stabilized by the intra-domain disulfide bonds. Overall, the IgM constant region has a “mushroom-like” structure, where the Cµ2-Cµ3 domains are a disk analogous to the head of the mushroom and the Cµ4tp domains protrude like a short stem.[38]
Function
IgM interacts with several other physiological molecules:
- IgM can bind complement component C1 and activate the classical pathway, leading to opsonization of antigens and cytolysis.
- IgM binds to the polyimmunoglobulin receptor (pIgR) in a process that brings IgM to mucosal surfaces, such as the gut lumen and into breast milk. This binding depends on J chain.[39]
- Two other Fc receptors that bind IgM—Fcα/µ-R and Fcµ-R -- have been detected. Fcα/µ-R, like pIgR, binds polymeric IgM and IgA. Fcα/µ-R can mediate endocytosis, and its expression in the gut suggests a role in mucosal immunity. Fcµ-R (formerly known as Toso/Faim3) binds IgM exclusively and can mediate cellular uptake of IgM-conjugated antigen.[40] Inactivation of the corresponding genes in knock-out mice produces a phenotype, but the physiological functions of these receptors are still uncertain[41]
Regulation of the immune response
Specific immunoglobulins that are injected into animals together with their antigen can influence the antibody response to this same antigen.[42] Endogenous antibodies produced after a primary immunization can also affect the antibody response to a booster immunization, suggesting that similar effects occur during physiological conditions. The ”regulatory” effects can be either positive or negative. That is, depending on the type of antigen and the isotype of the antibody, the effect can be suppression or enhancement of the antibody response. Such effects are well illustrated by experiments involving immunization with xenogenic (foreign) erythrocytes (red cells). For example, when IgG is administered together with xenogenic erythrocytes, this combination causes almost complete suppression of the erythrocyte-specific antibody response. This effect is used clinically to prevent Rh-negative mothers from becoming immunized against fetal Rh-positive erythrocytes, and its use has dramatically decreased the incidence of hemolytic disease in newborns.[43] In contrast to the effect of IgG, antigen-specific IgM can greatly enhance the antibody response, especially in the case of large antigens.[44] Thus, when IgM specific for erythrocytes is injected into animals (including humans) together with erythrocytes, a much stronger antibody response to the erythrocytes is induced than when erythrocytes are administered alone. Several lines of evidence indicate that the ability of IgM to activate complement is required for its enhancing effect. That is, IgM-mediated enhancement does not occur in animals that have been depleted for complement component C3, nor in mutant animals lacking complement receptors 1 and 2. Similarly, mutant IgM that cannot activate complement does not enhance the immune response. A possible explanation for IgM-mediated enhancement is that B lymphocytes capture IgM-antigen-complement complexes and transport the complexes into areas in the spleen where efficient immune responses are generated. Because IgM is produced early in an immune response, this might be important in the initiation of antibody responses.
Synthesis
In germ-line cells (sperm and ova) the genes that will eventually encode immunoglobulins are not in a functional form (see V(D)J recombination). In the case of the heavy chain, three germ-line segments denoted V, D and J are ligated together and adjoined to the DNA encoding the µ heavy chain constant region. Early in ontogeny, B cells express both the µ and the δ heavy chains; co-expression of these two heavy chains, each bearing the same V domain depends on alternative splicing and alternative poly-A addition sites. The expression of the other isotypes (γ, ε and α) is affected by another type of DNA rearrangement, a process called Immunoglobulin class switching.[45]
Clinical significance
IgM is the first immunoglobulin expressed in the human fetus (around 20 weeks)[46] and phylogenetically the earliest antibody to develop.[47]
IgM antibodies appear early in the course of an infection and usually reappear, to a lesser extent, after further exposure. IgM antibodies do not pass across the human placenta (only isotype IgG).
These two biological properties of IgM make it useful in the diagnosis of infectious diseases. Demonstrating IgM antibodies in a patient's serum indicates recent infection, or in a neonate's serum indicates intrauterine infection (e.g. congenital rubella syndrome).
The development of anti-donor IgM after organ transplantation is not associated with graft rejection but it may have a protective effect.[48]
IgM in normal serum is often found to bind to specific antigens, even in the absence of prior immunization.[49] For this reason, IgM has sometimes been called a "natural antibody". This phenomenon is probably due to the high avidity of IgM that allows it to bind detectably even to weakly cross-reacting antigens that are naturally occurring. For example, the IgM antibodies that bind to the red blood cell A and B antigens might be formed in early life as a result of exposure to A- and B-like substances that are present in bacteria or perhaps also in plant materials.
IgM antibodies are mainly responsible for the clumping (agglutination) of red blood cells if the recipient of a blood transfusion receives blood that is not compatible with their blood type.
A mutation of the mu chain causes autosomal recessive agammaglobulinemia.[50]
The presence of IgM or, rarely, IgG is one of the obligate criteria for a diagnosis of Schnitzler's syndrome.[51][52]
References
- "Immunoglobulin M". The American Heritage Dictionary of the English Language (Fourth ed.). Houghton Mifflin Company. 2004. ISBN 978-0618082308.
- Alberts, B.; Johnson, A.; Lewis, J.; Walter, P.; Raff, M.; Roberts, K. (2002). "Chapter 24". Molecular Biology of the Cell (4th ed.). Routledge. ISBN 978-0-8153-3288-6.
- Capolunghi, F.; Rosado, M. M.; Sinibaldi, M.; Aranburu, A.; Carsetti, R. (2013). "Why do we need IgM memory B cells?". Immunology Letters. 152 (2): 114–20. doi:10.1016/j.imlet.2013.04.007. PMID 23660557.
- Williams, N.; O'Connell, P. R. (2008). "Chapter 62". Bailey & Love's Short Practice of Surgery (25th ed.). CRC Press. p. 1102. ISBN 9780340939321.
- Heidelberger, M.; Pedersen, K. O. (1937). "The molecular weight of antibodies". Journal of Experimental Medicine. 65 (3): 393–414. doi:10.1084/jem.65.3.393. PMC 2133497. PMID 19870608.
- Kabat, E. A. (1939). "The molecular weight of antibodies". Journal of Experimental Medicine. 69 (1): 103–118. doi:10.1084/jem.69.1.103. PMC 2133729. PMID 19870830.
- Waldenström, J. (1943). "Incipient myelomatisis or "essential" hyoerglobulinemis with fibrinogenopenia—a new syndrome?". Acta Medica Scandinavica. 142 (3–4): 216–247. doi:10.1111/j.0954-6820.1944.tb03955.x.
- Potter, M. (2007). The early history of plasma cell tumors in mice, 1954-1976. Advances in Cancer Research. Vol. 98. pp. 17–51. doi:10.1016/S0065-230X(06)98002-6. ISBN 9780123738967. PMID 17433907.
- Monica, T. J.; Williams, S. B.; Goochee, C. F.; Maiorella, B. L. (1995). "Characterization of the glycosylation of a human IgM produced by a human-mouse hybridoma". Glycobiology. 5 (2): 175–185. doi:10.1093/glycob/5.2.175. PMID 7780192.
- Heyman, B.; Shulman, M. J. (2016). "Structure, Function, and Production of Immunoglobulin M (IgM)". In Ratcliffe, M. (ed.). Encyclopedia of Immunobiology. Vol. 2. Elsevier. pp. 1–14. doi:10.1016/B978-0-12-374279-7.05001-3. ISBN 978-0-12-374279-7.
- Beale, D.; Feinstein, A. (1969). "Studies on the Reduction of a Human 19S Immunoglobulin M". Biochemical Journal. 112 (2): 187–194. doi:10.1042/bj1120187. PMC 1187691. PMID 4979347.
- Milstein, C.P.; et al. (1975). "Interchain disulfide bridges of mouse Immunoglobulin M". Biochemical Journal. 151 (3): 615–624. doi:10.1042/bj1510615. PMC 1172409. PMID 766753.
- Halpern, M. S.; Koshland, M. E. (1970). "Novel subunit of secretory IgA". Nature. 228 (5278): 1276–1278. Bibcode:1970Natur.228.1276H. doi:10.1038/2281276a0. PMID 5530654. S2CID 36189290.
- Mestecky, J.; Zikin, J.; Butler, W. T. (1971). "Immunoglobulin M an secretory immunoglobulin A: presence of common polypeptide chain different from light chains". Science. 171 (3976): 1163–1165. Bibcode:1971Sci...171.1163M. doi:10.1126/science.171.3976.1163. PMID 5544873. S2CID 6834561.
- Frutiger, S.; et al. (1992). "Disulfide bond assignment in human J chain and its covalent pairing with immunoglobulin M". Biochemistry. 31 (50): 12643–12647. doi:10.1021/bi00165a014. PMID 1472500.
- Johansen, F. E.; Braathen, R.; Brandtzaeg, P. (2000). "Role of J chain in secretory immunoglobulin formation". Scandinavian Journal of Immunology. 52 (3): 240–8. doi:10.1046/j.1365-3083.2000.00790.x. PMID 10972899.
- Sørensen, V.; et al. (2000). "Structural requirements for incorporation of J chain into human IgM and IgA". International Immunology. 12 (1): 19–27. doi:10.1093/intimm/12.1.19. PMID 10607746.
- Cattaneo, A.; Neuberger, M. S. (1987). "Polymeric immunoglobulin M is secreted by transfectants of non-lymphoid cells in the absence of immunoglobulin J chain". The EMBO Journal. 6 (9): 2753–2758. doi:10.1002/j.1460-2075.1987.tb02569.x. PMC 553699. PMID 3119328.
- Fazel, S.; Wiersma, E. J.; Shulman, M. J. (1997). "Interplay of J chain and disulfide bonding in assembly of polymeric IgM". International Immunology. 9 (8): 1149–1158. doi:10.1093/intimm/9.8.1149. PMID 9263012.
- Parkhouse, R.; Askonas, B. A.; Dourmashkin, R. R. (1970). "Electron microscopic studies of mouse immunoglobulin M; structure and reconstitution following reduction". Immunology. 18 (4): 575–584. PMC 1455497. PMID 5421036.
- Schwager, J.; Hadji-Azlmi, I. (1984). "Mitogen-induced B-cell differentiation in Xenopus laevis". Differentiation. 27 (3): 182–188. doi:10.1111/j.1432-0436.1984.tb01426.x. PMID 6334001.
- Fillatreau, S.; et al. (2013). "The astonishing diversity of Ig classes and B cell repertoires in teleost fish". Frontiers in Immunology. 4: 1–14. doi:10.3389/fimmu.2013.00028. PMC 3570791. PMID 23408183.
- Getahun, A.; et al. (1999). "Influence of the μ-chain C-terminal sequence on polymerization of immunoglobulin M". Immunology. 97 (3): 408–413. doi:10.1046/j.1365-2567.1999.00797.x. PMC 2326861. PMID 10447761.
- Dolder, F. (1971). "Occurrence, Isolation and Interchain Bridges of Natural 7-S Immunoglobulin M in Human Serum". Biochimica et Biophysica Acta. 236 (3): 675–685. PMID 4997811.
- Eskeland, T.; Christensen, T. B. (1975). "IgM molecules with and without J chain in serum and after purification, studied by ultracentrifugation, electrophoresis, and electron microsopy". Scandinavian Journal of Immunology. 4 (3): 217–228. doi:10.1111/j.1365-3083.1975.tb02620.x. PMID 807966. S2CID 8246173.
- Davis, A. C.; Roux, K. H.; Shulman, M. J. (1988). "On the structure of polymeric IgM". European Journal of Immunology. 18 (7): 1001–1008. doi:10.1002/eji.1830180705. PMID 3136022. S2CID 34679165.
- Davis, A. C.; et al. (1989). "Intermolecular disulfide bonding in IgM: effects of replacing cysteine residues in the µ heavy chain". The EMBO Journal. 8 (9): 2519–2526. doi:10.1002/j.1460-2075.1989.tb08389.x. PMC 401247. PMID 2511005.
- Collins, C.; Tsui, F. W.; Shulman, M. J. (2002). "Differential activation of human and guinea pig complement by pentameric and hexameric IgM". European Journal of Immunology. 32 (6): 1802–1810. doi:10.1002/1521-4141(200206)32:6<1802::AID-IMMU1802>3.0.CO;2-C. PMID 12115664.
- Davis, A. C.; et al. (1989). "Mutations of the mouse m H chain which prevent polymer assembly". Journal of Immunology. 43 (4): 1352–1357. PMID 2501393.
- Smith, R. I. F.; Coloma, M. J.; Morrison, S. L. (1995). "Addition of a mu-tailpiece to IgG results in polymeric antibodies with enhanced effector functions including complement-mediated cytolysis by IgG4". Journal of Immunology. 154 (5): 2226–2236. PMID 7868896.
- Sørensen, V.; et al. (1996). "Effect of the IgM and IgA secretory tailpieces on polymerization and secretion of IgM and IgG". Journal of Immunology. 156 (8): 2858–2865. PMID 8609405.
- Smith, R.; Morrison, S. L. (1994). "Recombinant polymeric IgG: An approach to engineering more potent antibodies". Nature Biotechnology. 12 (7): 683–688. doi:10.1038/nbt0794-683. PMID 7764912. S2CID 19055205.
- Wiersma, E. J.; Shulman, M. J. (1995). "Assembly of IgM: role of disulfide bonding and noncovalent interactions". Journal of Immunology. 154 (10): 5265–5272. PMID 7730630.
- Chapuis, R. M.; Koshland, M. E. (1974). "Mechanism of IgM polymerization". Proc. Natl. Acad. Sci. U.S.A. 71 (3): 657–661. Bibcode:1974PNAS...71..657C. doi:10.1073/pnas.71.3.657. PMC 388071. PMID 4207070.
- Mihaesco, C.; Mihaesco, E.; Metzger, H. (1973). "Variable J-chain content in human IgM". FEBS Letters. 37 (2): 303–306. doi:10.1016/0014-5793(73)80483-1. PMID 4202824. S2CID 41601548.
- Brandtzaeg, P. (1976). "Complex formation between secretory component and human immunoglobulin related to their content of J chain". Scandinavian Journal of Immunology. 5 (4): 411–419. doi:10.1111/j.1365-3083.1976.tb00295.x. PMID 821140. S2CID 39847718.
- Grubb, A. O. (1978). "Quantitation of J chain in human biological fluids by a simple immunochemical procedure". Acta Medica Scandinavica. 204 (1–6): 453–465. doi:10.1111/j.0954-6820.1978.tb08473.x. PMID 104551.
- Müller, R.; et al. (2013). "High-resolutiion structures of the IgM Fc domainsreveal principles of its hexamer formation". Proc. Natl. Acad. Sci. U.S.A. 110 (25): 10183–10188. Bibcode:2013PNAS..11010183M. doi:10.1073/pnas.1300547110. PMC 3690842. PMID 23733956.
- Johansen, F. E.; Braathen, R.; Brandtzaeg, P. (2000). "Role of J chain in secretory immunoglobulin formation". Scandinavian Journal of Immunology. 52 (3): 240–8. doi:10.1046/j.1365-3083.2000.00790.x. PMID 10972899.
- Shima, H.; et al. (2010). "Identification of TOSO/FAIM3 as an Fc receptor for IgM". Int. Immunol. 22 (3): 149–56. doi:10.1093/intimm/dxp121. PMID 20042454.
- Ouchida, R.; et al. (2012). "Critical role of the IgM Fc receptor in IgM homeostasis, B-cell survival, and humoral immune responses". Proc. Natl. Acad. Sci. U.S.A. 109 (40): E2699–706. Bibcode:2012PNAS..109E2699O. doi:10.1073/pnas.1210706109. PMC 3479561. PMID 22988094.
- Heyman, B (2013). Antibody mediated regulation of humoral immunity. in Nimmerjahn, F. (ed.) Molecular and cellular mechanisms of antibody activity: Springer.
- Urbaniak, S. J. & Greiss, M. A. (2000). "RhD haemolytic disease of the fetus and the newborn". Blood Rev. 14 (1): 33–61. doi:10.1054/blre.1999.0123. PMID 10805260.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Sörman A, Zhang L, Ding Z, Heyman, B. (2014). "How antibodies use complement to regulate antibody responses". Mol. Immunol. 61 (2): 79–88. doi:10.1016/j.molimm.2014.06.010. PMID 25001046.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Murphy, K.; Weaver, C. (2016). Janeway's Immunobiology. New York, NY: Garland Science/Taylor and Francis. p. 195. ISBN 9780815345053.
- van Furth, R.; Schuit, H. R.; Hijmans, W. (1965). "The immunological development of the human fetus". Journal of Experimental Medicine. 122 (6): 1173–88. doi:10.1084/jem.122.6.1173. PMC 2138097. PMID 4159036.
- Review of Medical Physiology by William Francis Ganong
- McAlister, C. C.; et al. (2004). "Protective anti-donor IgM production after crossmatch positive liver-kidney transplantation". Liver Transplantation. 10 (2): 315–9. doi:10.1002/lt.20062. PMID 14762873.
- Jayasekera, J. P.; Moseman, E. A.; Carroll, M. C. (2007). "Natural antibody and complement mediate neutralization of influenza virus in the absence of prior immunity". Journal of Virology. 81 (7): 3487–94. doi:10.1128/JVI.02128-06. PMC 1866020. PMID 17202212.
- "OMIM Entry - # 601495 - AGAMMAGLOBULINEMIA 1, AUTOSOMAL RECESSIVE; AGM1". omim.org. Retrieved 2021-03-25.
- Gusdorf, L.; Lipsker, D. (17 July 2017). "Schnitzler Syndrome: a Review". Current Rheumatology Reports. 19 (8): 46. doi:10.1007/s11926-017-0673-5. PMID 28718061. S2CID 13780498.
- Simon, A.; Asli, B.; Braun-Falco, M.; Koning, H. De; Fermand, J.-P.; Grattan, C.; Krause, K.; Lachmann, H.; Lenormand, C.; Martinez-Taboada, V.; Maurer, M.; Peters, M.; Rizzi, R.; Rongioletti, F.; Ruzicka, T.; Schnitzler, L.; Schubert, B.; Sibilia, J.; Lipsker, D. (2013). "Schnitzler's syndrome: diagnosis, treatment, and follow-up". Allergy. 68 (5): 562–568. doi:10.1111/all.12129. PMID 23480774. S2CID 12831354.
External links
- Immunoglobulin+M at the US National Library of Medicine Medical Subject Headings (MeSH)
- Immunoglobulin M Deficiency Reference from Medscape.com