Macroevolution
Macroevolution usually means the evolution of large-scale structures and traits that go significantly beyond the intraspecific variation found in microevolution (including speciation).[1][2][3] In other words, macroevolution is the evolution of taxa above the species level (genera, families, orders, etc.).[4]
| Part of a series on |
| Evolutionary biology |
|---|
 |
|
Macroevolution is often thought to require the evolution of completely new structures such as entirely new organs. However, fundamentally novel structures are not necessary for dramatic evolutionary change. For instance, the evolution of mammal diversity in the past 100 million years has not required any major innovation.[5] All of this diversity can be explained by modification of existing organs.
In most cases, macroevolution cannot be observed directly because numerous mutations are required for large-scale changes. Hence specific methods are required to study and prove it. However, in rare cases single mutations can cause dramatic change and thus serve as models for macro-evolutionary processes.
Origin and changing meaning of the term
Philiptschenko[4] distinguished between microevolution and macroevolution because he rejected natural selection in the sense of Darwin[6] as an explanation for larger evolutionary transitions that give rise to taxa above the species level in the Linnean taxonomy. Accordingly, he restricted Darwinian "microevolution" to evolutionary changes within the boundary of given species that may lead to different races or subspecies at the most. By contrast, he referred "macroevolution" to major evolutionary changes that correspond to taxonomic differences above the species level, which in his opinion would require evolutionary processes different from natural selection. An explanatory model for macroevolution in this sense was the "hopeful monster" concept of geneticist Richard Goldschmidt, who suggested saltational evolutionary changes either due to mutations that affect the rates of developmental processes[7] or due to alterations in the chromosomal pattern.[8] Particularly the latter idea was widely rejected by the modern synthesis, but the hopeful monster concept based on evo-devo explanations found a moderate revival in recent times.[9][10] Occasionally such dramatic changes can lead to novel features that survive.
As an alternative to saltational evolution, Dobzhansky[11] suggested that the difference between macroevolution and microevolution reflects essentially a difference in time-scales, and that macroevolutionary changes were simply the sum of microevolutionary changes over geologic time. This view became broadly accepted, and accordingly, the term macroevolution has been used widely as a neutral label for the study of evolutionary changes that take place over a very large time-scale.[12]
Macroevolutionary processes
Species selection
"Species selection operates on variation provided by the largely random process of speciation and favors species that speciate at high rates or survive for long periods and therefore tend to leave many daughter species."[1] Species selection comprises (a) effect-macroevolution, where organism-level traits (aggregate traits) affect speciation and extinction rates (Stanley's original concept), and (b) strict-sense species selection, where species-level traits (e.g. geographical range) affect speciation and extinction rates.[13] It has been argued that effect macroevolution is reducible to microevolution because both operate through selection on organismic traits,[14] but Grantham[15] demonstrated that effect macroevolution can oppose selection at the organismic level and is therefore not reducible microevolution. Cases in which selection on the same trait has opposing effects at the organismic and the species level have been made in the context of sexual selection,[16][17][18] which increases individual fitness but may also increase the extinction risk of the species.
Evolution of new organs and tissues
One of the main questions in evolutionary biology is how fundamentally new structures evolve, such as new organs. As can be seen in vertebrate evolution, most "new" organs are actually not new—they are still modifications of previously existing organs. Examples are wings (modified limbs), feathers (modified reptile scales),[19] lungs (modified swim bladders, e.g. found in fish),[20][21] or even the heart (a muscularized segment of a vein).[22]
The same concept applies to the evolution of "novel" tissues. Even fundamental tissues such as bone can evolve from combining existing proteins (collagen) with calcium phosphate (specifically, hydroxy-apatite). This probably happened when certain cells that make collagen also accumulated calcium phosphate to get a proto-bone cell.[23]
Molecular macroevolution
Microevolution is facilitated by mutations, the vast majority of which have no or very small effects on gene or protein function. For instance, the activity of an enzyme may be slightly changed or the stability of a protein slightly altered. However, occasionally mutations can dramatically change the structure and functions of protein. This may be called "molecular macroevolution".
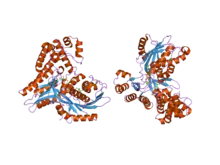
Protein function. There are countless cases in which protein function is dramatically altered by mutations. For instance, a mutation in acetaldehyde dehydrogenase (EC:1.2.1.10) can change it to a 4-hydroxy-2-oxopentanoate pyruvate lyase (EC:4.1.3.39), i.e., a mutation that changes an enzyme from one to another EC class.[24] Another example is the conversion of a yeast galactokinase (Gal1) to a transcription factor (Gal3) which can be achieved by an insertion of only two amino acids.[25]
While some mutations may not change the molecular function of a protein significantly, their biological function may be dramatically changed. For instance, most brain receptors recognize specific neurotransmitters, but that specificity can easily be changed by mutations. This has been shown by acetylcholine receptors that can be changed to serotonin or glycine receptors which actually have very different functions. Their similar gene structure also indicates that they must have arisen from gene duplications.[26]
Protein structure. Although protein structures are highly conserved, sometimes one or a few mutations can dramatically change a protein. For instance, an IgG-binding, 4+ fold can be transformed into an albumin-binding, 3-α fold via a single amino-acid mutation. This example also shows that such a transition can happen with neither function nor native structure being completely lost.[27] In other words, even when multiple mutations are required to convert one protein or structure into another, the structure and function is at least partially retained in the intermediary sequences.
Examples
Stanley's rule
Macroevolution is driven by differences between species in origination and extinction rates. Remarkably, these two factors are generally positively correlated: taxa that have typically high diversification rates have also high extinction rates. This observation has been described first by Steven Stanley, who attributed it to a variety of ecological factors.[28] Yet, a positive correlation of origination and extinction rates is also a prediction of the Red Queen hypothesis, which postulates that evolutionary progress (increase in fitness) of any given species causes a decrease in fitness of other species, ultimately driving to extinction those species that do not adapt rapidly enough.[29] High rates of origination must therefore correlate with high rates of extinction.[3] Stanley's rule, which applies to almost all taxa and geologic ages, is therefore a strong indication for a dominant role of biotic interactions in macroevolution.
"Macromutations": Single mutations leading to dramatic change
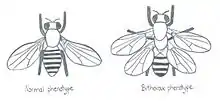
While the vast majority of mutations are inconsequential, some can have a dramatic effect on morphology or other features of an organism. One of the best studied cases of a single mutation that leads to massive structural change is the Ultrabithorax mutation in fruit flies. The mutation duplicates the wings of a fly to make it look like a dragonfly which represents a different order of insect.
Evolution of multicellularity
The evolution of multicellular organisms is one of the major breakthroughs in evolution. The first step of converting a unicellular organism into a metazoan (a multicellular organism) is to allow cells to attach to each other. This can be achieved by one or a few mutations. In fact, many bacteria form multicellular assemblies, e.g. cyanobacteria or myxobacteria. Another species of bacteria, Jeongeupia sacculi, form well-ordered sheets of cells, which ultimately develop into a bulbous structure.[30][31] Similarly, unicellular yeast cells can become multicellular by a single mutation in the ACE2 gene, which causes the cells to form a branched multicellular form.[32]
Evolution of bat wings
The wings of bats have the same structural elements (bones) as any other five-fingered mammal (see periodicity in limb development). However, the finger bones in bats are dramatically elongated, so the question is how these bones became so long. It has been shown that certain growth factors such as bone morphogenetic proteins (specifically Bmp2) is over expressed so that it stimulates an elongation of certain bones. Genetic changes in the bat genome identified the changes that lead to this phenotype and it has been recapitulated in mice: when specific bat DNA is inserted in the mouse genome, recapitulating these mutations, the bones of mice grow longer.[33]
Limbloss in lizards and snakes
.jpg.webp)
Snakes evolved from lizards. Phylogenetic analysis shows that snakes are actually nested within the phylogenetic tree of lizards, demonstrating that they have a common ancestor.[34] This split happened about 180 million years ago and several intermediary fossils are known to document the origin. In fact, limbs have been lost in numerous clades of reptiles, and there are cases of recent limb loss. For instance, the skink genus Lerista has lost limbs in multiple cases, with all possible intermediary steps, that is, there are species which have fully developed limbs, shorter limbs with 5, 4, 3, 2, 1 or no toes at all.[35]
Human evolution
While human evolution from their primate ancestors did not require massive morphological changes, our brain has sufficiently changed to allow human consciousness and intelligence. While the latter involves relatively minor morphological changes it did result in dramatic changes to brain function.[36] Thus, macroevolution does not have to be morphological, it can also be functional.
Evolution of viviparity in lizards

Most lizards are egg-laying and thus need an environment that is warm enough to incubate their eggs. However, some species have evolved viviparity, that is, they give birth to live young, as almost all mammals do. In several clades of lizards, egg-laying (oviparous) species have evolved into live-bearing ones, apparently with very little genetic change. For instance, a European common lizard, Zootoca vivipara, is vivparous throughout most of its range, but oviparous in the extreme southwest portion.[37][38] That is, within a single species, a radical change in reproductive behavior has happened. Similar cases are known from South American lizards of the genus Liolaemus which have egg-laying species at lower altitudes, but closely related viviparous species at higher altitudes, suggesting that the switch from oviparous to viviparous reproduction does not require many genetic changes.[39]
Criticism
Evolution as a biological process is now broadly accepted by the scientific community, but also by circles who were formerly opposed to it, such as the Catholic church.[40] However, even people who accept evolution on a "micro" level often remain skeptical about "macroevolution". Below are some arguments critics have brought forward against macroevolution as well as counter-arguments to these objections.

Lack of observation and missing links. While microevolution can be observed and measured in living organisms (e.g. as genetic variation or selection within populations), critics argue that macroevolution cannot be observed, given that it occurs over millions of years. Long-term changes as "observed" through the fossil record are usually spotty so that links between different species are often missing. This argument has been refuted by the fact that many transitional forms have indeed been found (e.g. Archaeopteryx and living examples above). More importantly, transitions have been meticulously documented in the genomes of organisms (e.g. all mutations are known that happened in human evolution since our lineage split from that leading to chimpanzees).[41][42] Finally, historical processes can be recapitulated in the laboratory and thus observed using experiments (e.g., see evolution of bat wings, above).
Irreducible complexity. Large transitions in evolution are said to be impossible because many structures in biology are "irreducibly complex".[43] The most cited example is the bacterial flagellum, a multiprotein complex that, allegedly, cannot evolve from simpler structures because any simpler ancestor would be non-functional. However, this claim has been debunked numerous times: many proteins of the flagellar apparatus can be deleted without loss of function, hence the structure is certainly not "irreducibly complex".[44] In addition, there is convincing evidence that flagella have evolved from much simpler structures similar to type III secretion systems.[45]
Research topics
Subjects studied within macroevolution include:[46]
- Adaptive radiations such as the Cambrian Explosion.
- Changes in biodiversity through time.
- Genome evolution, like horizontal gene transfer, genome fusions in endosymbioses, and adaptive changes in genome size.
- Mass extinctions.
- Estimating diversification rates, including rates of speciation and extinction.
- The debate between punctuated equilibrium and gradualism.
- The role of development in shaping evolution, particularly such topics as heterochrony and phenotypic plasticity.
See also
- Extinction event
- Interspecific competition
- Microevolution
- Molecular evolution
- Punctuated equilibrium
- Red Queen hypothesis
- Speciation
- Unit of selection
References
- Stanley, S. M. (1 February 1975). "A theory of evolution above the species level". Proceedings of the National Academy of Sciences. 72 (2): 646–50. Bibcode:1975PNAS...72..646S. doi:10.1073/pnas.72.2.646. ISSN 0027-8424. PMC 432371. PMID 1054846.
- Gould, Stephen Jay (2002). The structure of evolutionary theory. Cambridge, Mass.: Belknap Press of Harvard University Press. ISBN 0-674-00613-5. OCLC 47869352.
- Hautmann, Michael (2020). "What is macroevolution?". Palaeontology. 63 (1): 1–11. doi:10.1111/pala.12465. ISSN 0031-0239.
- Philiptschenko, J. (1927). Variabilität und Variation. Berlin: Borntraeger.
- Meredith, R. W.; Janecka, J. E.; Gatesy, J.; Ryder, O. A.; Fisher, C. A.; Teeling, E. C.; Goodbla, A.; Eizirik, E.; Simao, T. L. L.; Stadler, T.; Rabosky, D. L.; Honeycutt, R. L.; Flynn, J. J.; Ingram, C. M.; Steiner, C. (28 October 2011). "Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification". Science. 334 (6055): 521–524. doi:10.1126/science.1211028. ISSN 0036-8075.
- Darwin, C. (1859). On the origin of species by means of natural selection. London: John Murray.
- Goldschmidt, R. (1933). "Some aspects of evolution". Science. 78 (2033): 539–547. Bibcode:1933Sci....78..539G. doi:10.1126/science.78.2033.539. PMID 17811930.
- Goldschmidt, R. (1940). The material basis of evolution. Yale University Press.
- Theißen, Günter (March 2009). "Saltational evolution: hopeful monsters are here to stay". Theory in Biosciences. 128 (1): 43–51. doi:10.1007/s12064-009-0058-z. ISSN 1431-7613. PMID 19224263. S2CID 4983539.
- Rieppel, Olivier (13 March 2017). Turtles as hopeful monsters : origins and evolution. Bloomington, Indiana. ISBN 978-0-253-02507-4. OCLC 962141060.
- Dobzhanski, T. (1937). Genetics and the origin of species. Columbia University Press.
- Dawkins, Richard, 1941- (1982). The extended phenotype : the gene as the unit of selection. Oxford [Oxfordshire]: Freeman. ISBN 0-7167-1358-6. OCLC 7652745.
{{cite book}}: CS1 maint: multiple names: authors list (link) - Jablonski, David (December 2008). "Species Selection: Theory and Data". Annual Review of Ecology, Evolution, and Systematics. 39 (1): 501–524. doi:10.1146/annurev.ecolsys.39.110707.173510. ISSN 1543-592X.
- Greenwood, P. H. (1979). "Macroevolution - myth or reality ?". Biological Journal of the Linnean Society. 12 (4): 293–304. doi:10.1111/j.1095-8312.1979.tb00061.x.
- Grantham, T A (November 1995). "Hierarchical Approaches to Macroevolution: Recent Work on Species Selection and the "Effect Hypothesis"". Annual Review of Ecology and Systematics. 26 (1): 301–321. doi:10.1146/annurev.es.26.110195.001505. ISSN 0066-4162.
- McLain, Denson K.; Moulton, Michael P.; Redfearn, Todd P. (October 1995). "Sexual Selection and the Risk of Extinction of Introduced Birds on Oceanic Islands". Oikos. 74 (1): 27. doi:10.2307/3545671. ISSN 0030-1299. JSTOR 3545671.
- Moen, R. A. "Antler growth and extinction of Irish elk". Evolutionary Ecology Research. 1: 235–249.
- Martins, Maria João Fernandes; Puckett, T. Markham; Lockwood, Rowan; Swaddle, John P.; Hunt, Gene (April 2018). "High male sexual investment as a driver of extinction in fossil ostracods". Nature. 556 (7701): 366–369. Bibcode:2018Natur.556..366M. doi:10.1038/s41586-018-0020-7. ISSN 0028-0836. PMID 29643505. S2CID 4925632.
- Wu, Ping; Yan, Jie; Lai, Yung-Chih; Ng, Chen Siang; Li, Ang; Jiang, Xueyuan; Elsey, Ruth M; Widelitz, Randall; Bajpai, Ruchi; Li, Wen-Hsiung; Chuong, Cheng-Ming (21 November 2017). "Multiple Regulatory Modules Are Required for Scale-to-Feather Conversion". Molecular Biology and Evolution. 35 (2): 417–430. doi:10.1093/molbev/msx295. ISSN 0737-4038. PMC 5850302. PMID 29177513.
- Brainerd, E. L. (1 December 1999). "New perspectives on the evolution of lung ventilation mechanisms in vertebrates". Experimental Biology Online. 4 (2): 1–28. doi:10.1007/s00898-999-0002-1. ISSN 1430-3418.
- Hoffman, M.; Taylor, B. E.; Harris, M. B. (1 April 2016). "Evolution of lung breathing from a lungless primitive vertebrate". Respiratory Physiology & Neurobiology. Physiology of respiratory networks of non-mammalian vertebrates. 224: 11–16. doi:10.1016/j.resp.2015.09.016. ISSN 1569-9048. PMC 5138057. PMID 26476056.
- Jensen, Bjarke; Wang, Tobias; Christoffels, Vincent M.; Moorman, Antoon F. M. (1 April 2013). "Evolution and development of the building plan of the vertebrate heart". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. Cardiomyocyte Biology: Cardiac Pathways of Differentiation, Metabolism and Contraction. 1833 (4): 783–794. doi:10.1016/j.bbamcr.2012.10.004. ISSN 0167-4889.
- Wagner, Darja Obradovic; Aspenberg, Per (1 August 2011). "Where did bone come from?". Acta Orthopaedica. 82 (4): 393–398. doi:10.3109/17453674.2011.588861. ISSN 1745-3674. PMC 3237026. PMID 21657973.
- Tyzack, Jonathan D; Furnham, Nicholas; Sillitoe, Ian; Orengo, Christine M; Thornton, Janet M (1 December 2017). "Understanding enzyme function evolution from a computational perspective". Current Opinion in Structural Biology. Protein–nucleic acid interactions • Catalysis and regulation. 47: 131–139. doi:10.1016/j.sbi.2017.08.003. ISSN 0959-440X.
- Platt, A.; Ross, H. C.; Hankin, S.; Reece, R. J. (28 March 2000). "The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase". Proceedings of the National Academy of Sciences of the United States of America. 97 (7): 3154–3159. doi:10.1073/pnas.97.7.3154. ISSN 0027-8424. PMC 16208. PMID 10737789.
- Uetz, Peter; Abdelatty, Fawzy; Villarroel, Alfredo; Rappold, Gudrun; Weiss, Birgit; Koenen, Michael (21 February 1994). "Organisation of the murine 5-HT 3 receptor gene and assignment tohuman chromosome 11". FEBS Letters. 339 (3): 302–306. doi:10.1016/0014-5793(94)80435-4.
- Alexander, Patrick A.; He, Yanan; Chen, Yihong; Orban, John; Bryan, Philip N. (15 December 2009). "A minimal sequence code for switching protein structure and function". Proceedings of the National Academy of Sciences. 106 (50): 21149–21154. doi:10.1073/pnas.0906408106. ISSN 0027-8424. PMC 2779201. PMID 19923431.
- Stanley, Steven M. (1979). Macroevolution, pattern and process. San Francisco: W.H. Freeman. ISBN 0-7167-1092-7. OCLC 5101557.
- Van Valen, L. (1973). "A new evolutionary law". Evolutionary Theory. 1: 1–30.
- Datta, Sayantan; Ratcliff, William C (11 October 2022). "Illuminating a new path to multicellularity". eLife. 11: e83296. doi:10.7554/eLife.83296. ISSN 2050-084X. PMC 9553208.
- Mizuno, Kouhei; Maree, Mais; Nagamura, Toshihiko; Koga, Akihiro; Hirayama, Satoru; Furukawa, Soichi; Tanaka, Kenji; Morikawa, Kazuya (11 October 2022). Goldstein, Raymond E; Weigel, Detlef (eds.). "Novel multicellular prokaryote discovered next to an underground stream". eLife. 11: e71920. doi:10.7554/eLife.71920. ISSN 2050-084X.
- Ratcliff, William C.; Fankhauser, Johnathon D.; Rogers, David W.; Greig, Duncan; Travisano, Michael (May 2015). "Origins of multicellular evolvability in snowflake yeast". Nature Communications. 6 (1): 6102. doi:10.1038/ncomms7102. ISSN 2041-1723. PMC 4309424. PMID 25600558.
- Sears, Karen E.; Behringer, Richard R.; Rasweiler, John J.; Niswander, Lee A. (25 April 2006). "Development of bat flight: Morphologic and molecular evolution of bat wing digits". Proceedings of the National Academy of Sciences. 103 (17): 6581–6586. doi:10.1073/pnas.0509716103. ISSN 0027-8424. PMC 1458926. PMID 16618938.
- Streicher, Jeffrey W.; Wiens, John J. (30 September 2017). "Phylogenomic analyses of more than 4000 nuclear loci resolve the origin of snakes among lizard families". Biology Letters. 13 (9): 20170393. doi:10.1098/rsbl.2017.0393. PMC 5627172. PMID 28904179.
- Skinner, Adam; Lee, Michael SY; Hutchinson, Mark N (2008). "Rapid and repeated limb loss in a clade of scincid lizards". BMC Evolutionary Biology. 8 (1): 310. doi:10.1186/1471-2148-8-310. ISSN 1471-2148. PMC 2596130. PMID 19014443.
- Serrelli, Emanuele; Gontier, Nathalie (2015). Macroevolution: explanation, interpretation and evidence. Cham. ISBN 978-3-319-15045-1. OCLC 903489046.
- Heulin, Benoît (1 May 1990). "Étude comparative de la membrane coquillère chez les souches ovipare et vivipare du lézard Lacerta vivipara". Canadian Journal of Zoology. 68 (5): 1015–1019. doi:10.1139/z90-147. ISSN 0008-4301.
- Arrayago, Maria-Jesus; Bea, Antonio; Heulin, Benoit (1996). "Hybridization Experiment between Oviparous and Viviparous Strains of Lacerta vivipara: A New Insight into the Evolution of Viviparity in Reptiles". Herpetologica. 52 (3): 333–342. ISSN 0018-0831.
- Ii, James A. Schulte; Macey, J. Robert; Espinoza, Robert E.; Larson, Allan (January 2000). "Phylogenetic relationships in the iguanid lizard genus Liolaemus: multiple origins of viviparous reproduction and evidence for recurring Andean vicariance and dispersal". Biological Journal of the Linnean Society. 69 (1): 75–102. doi:10.1111/j.1095-8312.2000.tb01670.x.
- Magazine, Smithsonian; Schultz, Colin. "The Pope Would Like You to Accept Evolution and the Big Bang". Smithsonian Magazine. Retrieved 18 September 2022.
- Kehrer-Sawatzki, Hildegard; Cooper, David N. (February 2007). "Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons". Human Mutation. 28 (2): 99–130. doi:10.1002/humu.20420.
- Varki, Ajit; Altheide, Tasha K. (1 December 2005). "Comparing the human and chimpanzee genomes: Searching for needles in a haystack". Genome Research. 15 (12): 1746–1758. doi:10.1101/gr.3737405. ISSN 1088-9051. PMID 16339373.
- Behe, Michael J. (2007). The edge of evolution : the search for the limits of Darwinism. New York: Free Press. ISBN 978-0-7432-9620-5. OCLC 136958644.
- Rajagopala, Seesandra V; Titz, Björn; Goll, Johannes; Parrish, Jodi R; Wohlbold, Katrin; McKevitt, Matthew T; Palzkill, Timothy; Mori, Hirotada; Finley, Russell L; Uetz, Peter (January 2007). "The protein network of bacterial motility". Molecular Systems Biology. 3 (1): 128. doi:10.1038/msb4100166. ISSN 1744-4292. PMC 1943423. PMID 17667950.
- Liu, Renyi; Ochman, Howard (24 April 2007). "Stepwise formation of the bacterial flagellar system". Proceedings of the National Academy of Sciences. 104 (17): 7116–7121. doi:10.1073/pnas.0700266104. ISSN 0027-8424.
- Grinin, L., Markov, A. V., Korotayev, A. Aromorphoses in Biological and Social Evolution: Some General Rules for Biological and Social Forms of Macroevolution / Social evolution & History, vol.8, num. 2, 2009
Further reading
- What is marcroevolution? (pdf) https://onlinelibrary.wiley.com/doi/full/10.1111/pala.12465
- AAAS, American Association for the Advancement of Science (16 February 2006). "Statement on the Teaching of Evolution" (PDF). aaas.org. Archived from the original (PDF) on 21 February 2006. Retrieved 14 January 2007.
- IAP, Interacademy Panel (21 June 2006). IAP Statement on the Teaching of Evolution (PDF). interacademies.net. Archived from the original (PDF) on 5 July 2006. Retrieved 14 January 2007.
- Myers, P.Z. (18 June 2006). "Ann Coulter: No Evidence for Evolution?". Pharyngula. ScienceBlogs. Archived from the original on 22 June 2006. Retrieved 12 September 2007.
- NSTA, National Science Teachers Association (2007). "An NSTA Evolution Q&A". Archived from the original on 2 February 2008. Retrieved 1 February 2008.
- Pinholster, Ginger (19 February 2006). "AAAS Denounces Anti-Evolution Laws as Hundreds of K-12 Teachers Convene for 'Front Line' Event". aaas.org. Retrieved 14 January 2007.
External links
- Introduction to macroevolution
- Macroevolution as the common descent of all life
- Macroevolution in the 21st century Macroevolution as an independent discipline.
- Macroevolution FAQ
