CD86
Cluster of Differentiation 86 (also known as CD86 and B7-2) is a protein constitutively expressed on dendritic cells, Langerhans cells, macrophages, B-cells (including memory B-cells), and on other antigen-presenting cells.[5] Along with CD80, CD86 provides costimulatory signals necessary for T cell activation and survival. Depending on the ligand bound, CD86 can signal for self-regulation and cell-cell association, or for attenuation of regulation and cell-cell disassociation.[6]
The CD86 gene encodes a type I membrane protein that is a member of the immunoglobulin superfamily.[7] Alternative splicing results in two transcript variants encoding different isoforms. Additional transcript variants have been described, but their full-length sequences have not been determined.[8]
Structure
CD86 belongs to the B7 family of the immunoglobulin superfamily.[9] It is a 70 kDa glycoprotein made up of 329 amino acids. Both CD80 and CD86 share a conserved amino acid motif that forms their ligand binding domain.[10] CD86 consists of Ig-like extracellular domains (one variable and one constant), a transmembrane region and a short cytoplasmic domain that is longer than that of CD80.[11][12] costimulatory ligands CD80 and CD86 can be found on professional antigen presenting cells such as monocytes, dendritic cells, and even activated B-cells. They can also be induced on other cell types, for example T cells.[13] CD86 expression is more abundant compared to CD80, and upon its activation is CD86 increased faster than CD80.[14]
At the protein level, CD86 shares 25% identity with CD80[15] and both are coded on human chromosome 3q13.33q21.[16]
Role in co-stimulation, T-cell activation and inhibition
CD86 and CD80 bind as ligands to costimulatory molecule CD28 on the surface of all naïve T cells,[17] and to the inhibitory receptor CTLA-4 (cytotoxic T-lymphocyte antigen-4, also known as CD152).[18][19] CD28 and CTLA-4 have important, but opposite roles in the stimulation of T cells. Binding to CD28 promotes T cell responses, while binding to CTLA-4 inhibits them.[20]
The interaction between CD86 (CD80) expressed on the surface of an antigen-presenting cell with CD28 on the surface of a mature, naive T-cell, is required for T-cell activation.[21] To become activated, lymphocyte must engage both antigen and costimulatory ligand on the same antigen-presenting cell. T cell receptor (TCR) interacts with major histocompatibility complex (MHC) class II molecules,[13] and this signalization must be accompanied by costimulatory signals, provided by a costimulatory ligand. These costimulatory signals are necessary to prevent anergy and are provided by the interaction between CD80/CD86 and CD28 costimulatory molecule.[22][23]
This protein interaction is also essential for T lymphocytes to receive the full activation signal, which in turn leads to T cell differentiation and division, production of interleukin 2 and clonal expansion.[9][22] Interaction between CD86 and CD28 activates mitogen-activated protein kinase and transcription factor nf-κB in the T-cell. These proteins up-regulate production of CD40L (used in B-cell activation), IL-21 and IL-21R (used for division/proliferation), and IL-2, among other cytokines.[21] The interaction also regulates self-tolerance by supporting the homeostatis of CD4+CD25+ Tregulatory cell, also known as Tregs.[9]
CTLA-4 is a coinhibitory molecule that is induced on activated T cells. Interaction between CTLA-4 and CD80/CD86 leads to delivery of negative signals into T cells and reduction of number of costimulatory molecules on the cell surface. It can also trigger a signaling pathway responsible for expression of enzyme IDO (indolamine-2,3-dioxygenase). This enzyme can metabolize amino acid tryptophan, which is an important component for successful proliferation and differentiation of T lymphocytes. IDO reduces the concentration of tryptophan in the environment, thereby suppressing the activation of conventional T cells, while also promoting the function of regulatory T cells.[24][25]
Both CD80 and CD86 bind CTLA-4 with higher affinity than CD28. This allows CTLA-4 to outcompete CD28 for CD80/CD86 binding.[23][26] Between CD80 and CD86, CD80 appears to have a higher affinity for both CTLA-4 and CD28 than CD86. This suggest that CD80 is more potent ligand than CD86,[15] but studies using CD80 and CD86 knockout mice have shown that CD86 is more important in T cell activation than CD80.[27]
Treg mediation
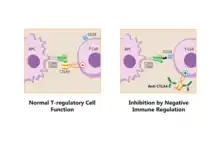
Pathways in the B7:CD28 family have key roles in the regulation of T cell activation and tolerance. Their negative second signals are responsible for downregulation of cell responses. For all these reasons are these pathways considered as therapeutic targets.[9]
Regulatory T cells produce CTLA-4. Due to its interaction with CD80/CD86, Tregs can compete with conventional T cells and block their costimulatory signals. Treg expression of CTLA-4 can effectively downregulate both CD80 and CD86 on APCs,[28] suppress the immune response and lead to increased anergy.[6] Since CTLA-4 binds to CD86 with higher affinity than CD28, the co-stimulation necessary for proper T-cell activation is also affected.[29] It was shown in a work from Sagurachi group that Treg cells were able to downregulate CD80 and CD86, but not CD40 or MHC class II on DC in a way that was adhesion dependent. Downregulation was blocked by anti-CTLA-4 antibody and was cancelled if Treg cells were CTLA-4 deficient.[30]
When bound to CTLA-4, CD86 can be removed from the surface of an APC and onto the Treg cell in a process called trogocytosis.[6] Blocking this process with anti-CTLA-4 antibodies is useful for a specific type of cancer immunotherapy called "Cancer therapy by inhibition of negative immune regulation".[31] Japanese immunologist Tasuku Honjo and American immunologist James P. Allison won the Nobel Prize in Physiology or Medicine in 2018 for their work on this topic.
Role in pathology
Roles of both CD80 and CD86 are studied in context of many pathologies. Selective inhibition of costimulatory inhibitors was examined in a model of allergic pulmonary inflammation and airway hyper-responsiveness (AHR).[32] Since initial host response to Staphylococcus aureus, especially the immune response based on T cells, is a contributing factor in the pathogenesis of acute pneumonia, role of the CD80/CD86 pathway in pathogenesis was investigated.[33] The costimulatory molecules were also investigated in context of Bronchial Astma,[34] Treg in cancer,[35] and immunotherapy.[36]
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000114013 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000022901 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ Lenschow DJ, Su GH, Zuckerman LA, Nabavi N, Jellis CL, Gray GS, et al. (December 1993). "Expression and functional significance of an additional ligand for CTLA-4". Proceedings of the National Academy of Sciences of the United States of America. 90 (23): 11054–8. Bibcode:1993PNAS...9011054L. doi:10.1073/pnas.90.23.11054. PMC 47920. PMID 7504292.
- 1 2 3 Ohue Y, Nishikawa H (July 2019). "Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target?". Cancer Science. 110 (7): 2080–2089. doi:10.1111/cas.14069. PMC 6609813. PMID 31102428.
- ↑ Chen C, Gault A, Shen L, Nabavi N (May 1994). "Molecular cloning and expression of early T cell costimulatory molecule-1 and its characterization as B7-2 molecule". Journal of Immunology. 152 (10): 4929–36. PMID 7513726.
- ↑ "Entrez Gene: CD86 CD86 molecule".
- 1 2 3 4 Greenwald RJ, Freeman GJ, Sharpe AH (2005). "The B7 family revisited". Annual Review of Immunology. 23: 515–48. doi:10.1146/annurev.immunol.23.021704.115611. PMID 15771580.
- ↑ Yu C, Sonnen AF, George R, Dessailly BH, Stagg LJ, Evans EJ, et al. (February 2011). "Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering". The Journal of Biological Chemistry. 286 (8): 6685–96. doi:10.1074/jbc.M110.182394. PMC 3057841. PMID 21156796.
- ↑ Freeman GJ, Borriello F, Hodes RJ, Reiser H, Hathcock KS, Laszlo G, et al. (November 1993). "Uncovering of functional alternative CTLA-4 counter-receptor in B7-deficient mice". Science. 262 (5135): 907–9. Bibcode:1993Sci...262..907F. doi:10.1126/science.7694362. PMID 7694362.
- ↑ Sharpe AH, Freeman GJ (February 2002). "The B7-CD28 superfamily". Nature Reviews. Immunology. 2 (2): 116–26. doi:10.1038/nri727. PMID 11910893. S2CID 205492817.
- 1 2 Murphy K, Weaver C, Janeway C (2017). Janeway's immunobiology (9th ed.). New York. ISBN 978-0-8153-4505-3. OCLC 933586700.
- ↑ Sansom DM (October 2000). "CD28, CTLA-4 and their ligands: who does what and to whom?". Immunology. 101 (2): 169–77. doi:10.1046/j.1365-2567.2000.00121.x. PMC 2327073. PMID 11012769.
- 1 2 Collins AV, Brodie DW, Gilbert RJ, Iaboni A, Manso-Sancho R, Walse B, et al. (August 2002). "The interaction properties of costimulatory molecules revisited". Immunity. 17 (2): 201–10. doi:10.1016/s1074-7613(02)00362-x. PMID 12196291.
- ↑ Mir MA (25 May 2015). Developing costimulatory molecules for immunotherapy of diseases. London. ISBN 978-0-12-802675-5. OCLC 910324332.
- ↑ Linsley PS, Brady W, Grosmaire L, Aruffo A, Damle NK, Ledbetter JA (March 1991). "Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation". The Journal of Experimental Medicine. 173 (3): 721–30. doi:10.1084/jem.173.3.721. PMC 2118836. PMID 1847722.
- ↑ Lim TS, Goh JK, Mortellaro A, Lim CT, Hämmerling GJ, Ricciardi-Castagnoli P (2012). "CD80 and CD86 differentially regulate mechanical interactions of T-cells with antigen-presenting dendritic cells and B-cells". PLOS ONE. 7 (9): e45185. Bibcode:2012PLoSO...745185L. doi:10.1371/journal.pone.0045185. PMC 3443229. PMID 23024807.
- ↑ Linsley PS, Brady W, Urnes M, Grosmaire LS, Damle NK, Ledbetter JA (September 1991). "CTLA-4 is a second receptor for the B cell activation antigen B7". The Journal of Experimental Medicine. 174 (3): 561–9. doi:10.1084/jem.174.3.561. PMC 2118936. PMID 1714933.
- ↑ Sansom DM, Manzotti CN, Zheng Y (June 2003). "What's the difference between CD80 and CD86?". Trends in Immunology. 24 (6): 314–9. doi:10.1016/s1471-4906(03)00111-x. PMID 12810107.
- 1 2 Dyck L, Mills KH (May 2017). "Immune checkpoints and their inhibition in cancer and infectious diseases". European Journal of Immunology. 47 (5): 765–779. doi:10.1002/eji.201646875. PMID 28393361.
- 1 2 Coyle AJ, Gutierrez-Ramos JC (March 2001). "The expanding B7 superfamily: increasing complexity in costimulatory signals regulating T cell function". Nature Immunology. 2 (3): 203–9. doi:10.1038/85251. PMID 11224518. S2CID 20542148.
- 1 2 Gause WC, Urban JF, Linsley P, Lu P (1995). "Role of B7 signaling in the differentiation of naive CD4+ T cells to effector interleukin-4-producing T helper cells". Immunologic Research. 14 (3): 176–88. doi:10.1007/BF02918215. PMID 8778208. S2CID 20098311.
- ↑ Chen L, Flies DB (April 2013). "Molecular mechanisms of T cell co-stimulation and co-inhibition". Nature Reviews. Immunology. 13 (4): 227–42. doi:10.1038/nri3405. PMC 3786574. PMID 23470321.
- ↑ Munn DH, Sharma MD, Mellor AL (April 2004). "Ligation of B7-1/B7-2 by human CD4+ T cells triggers indoleamine 2,3-dioxygenase activity in dendritic cells". Journal of Immunology. 172 (7): 4100–10. doi:10.4049/jimmunol.172.7.4100. PMID 15034022.
- ↑ Walker LS, Sansom DM (November 2011). "The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses". Nature Reviews. Immunology. 11 (12): 852–63. doi:10.1038/nri3108. PMID 22116087. S2CID 9617595.
- ↑ Borriello F, Sethna MP, Boyd SD, Schweitzer AN, Tivol EA, Jacoby D, et al. (March 1997). "B7-1 and B7-2 have overlapping, critical roles in immunoglobulin class switching and germinal center formation". Immunity. 6 (3): 303–13. doi:10.1016/s1074-7613(00)80333-7. PMID 9075931.
- ↑ Walker LS, Sansom DM (February 2015). "Confusing signals: recent progress in CTLA-4 biology". Trends in Immunology. 36 (2): 63–70. doi:10.1016/j.it.2014.12.001. PMC 4323153. PMID 25582039.
- ↑ Lightman SM, Utley A, Lee KP (2019-05-03). "Survival of Long-Lived Plasma Cells (LLPC): Piecing Together the Puzzle". Frontiers in Immunology. 10: 965. doi:10.3389/fimmu.2019.00965. PMC 6510054. PMID 31130955.
- ↑ Onishi Y, Fehervari Z, Yamaguchi T, Sakaguchi S (July 2008). "Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation". Proceedings of the National Academy of Sciences of the United States of America. 105 (29): 10113–8. Bibcode:2008PNAS..10510113O. doi:10.1073/pnas.0711106105. PMC 2481354. PMID 18635688.
- ↑ Chen R, Ganesan A, Okoye I, Arutyunova E, Elahi S, Lemieux MJ, Barakat K (March 2020). "Targeting B7-1 in immunotherapy". Medicinal Research Reviews. 40 (2): 654–682. doi:10.1002/med.21632. PMID 31448437. S2CID 201748060.
- ↑ Mark DA, Donovan CE, De Sanctis GT, Krinzman SJ, Kobzik L, Linsley PS, et al. (November 1998). "Both CD80 and CD86 co-stimulatory molecules regulate allergic pulmonary inflammation". International Immunology. 10 (11): 1647–55. doi:10.1093/intimm/10.11.1647. PMID 9846693.
- ↑ Parker D (July 2018). "CD80/CD86 signaling contributes to the proinflammatory response of Staphylococcus aureus in the airway". Cytokine. 107: 130–136. doi:10.1016/j.cyto.2018.01.016. PMC 5916031. PMID 29402722.
- ↑ Chen YQ, Shi HZ (January 2006). "CD28/CTLA-4--CD80/CD86 and ICOS--B7RP-1 costimulatory pathway in bronchial asthma". Allergy. 61 (1): 15–26. doi:10.1111/j.1398-9995.2006.01008.x. PMID 16364152. S2CID 23564785.
- ↑ Ohue Y, Nishikawa H (July 2019). "Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target?". Cancer Science. 110 (7): 2080–2089. doi:10.1111/cas.14069. PMC 6609813. PMID 31102428.
- ↑ Bourque J, Hawiger D (2018). "Immunomodulatory Bonds of the Partnership between Dendritic Cells and T Cells". Critical Reviews in Immunology. 38 (5): 379–401. doi:10.1615/CritRevImmunol.2018026790. PMC 6380512. PMID 30792568.
External links
Further reading
- Davila S, Froeling FE, Tan A, Bonnard C, Boland GJ, Snippe H, et al. (April 2010). "New genetic associations detected in a host response study to hepatitis B vaccine". Genes and Immunity. 11 (3): 232–8. doi:10.1038/gene.2010.1. PMID 20237496.
- Csillag A, Boldogh I, Pazmandi K, Magyarics Z, Gogolak P, Sur S, et al. (March 2010). "Pollen-induced oxidative stress influences both innate and adaptive immune responses via altering dendritic cell functions". Journal of Immunology. 184 (5): 2377–85. doi:10.4049/jimmunol.0803938. PMC 3028537. PMID 20118277.
- Bossé Y, Lemire M, Poon AH, Daley D, He JQ, Sandford A, et al. (October 2009). "Asthma and genes encoding components of the vitamin D pathway". Respiratory Research. 10: 98. doi:10.1186/1465-9921-10-98. PMC 2779188. PMID 19852851.
- Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH, et al. (May 2010). "Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus". The Journal of Infectious Diseases. 201 (9): 1371–80. doi:10.1086/651606. PMC 2853721. PMID 20331378.
- Bugeon L, Dallman MJ (October 2000). "Costimulation of T cells". American Journal of Respiratory and Critical Care Medicine. 162 (4 Pt 2): S164-8. doi:10.1164/ajrccm.162.supplement_3.15tac5. PMID 11029388.
- Pan XM, Gao LB, Liang WB, Liu Y, Zhu Y, Tang M, et al. (July 2010). "CD86 +1057 G/A polymorphism and the risk of colorectal cancer". DNA and Cell Biology. 29 (7): 381–6. doi:10.1089/dna.2009.1003. PMID 20380573.
- Dalla-Costa R, Pincerati MR, Beltrame MH, Malheiros D, Petzl-Erler ML (August 2010). "Polymorphisms in the 2q33 and 3q21 chromosome regions including T-cell coreceptor and ligand genes may influence susceptibility to pemphigus foliaceus". Human Immunology. 71 (8): 809–17. doi:10.1016/j.humimm.2010.04.001. PMID 20433886.
- Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, et al. (November 2009). "Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip". American Journal of Human Genetics. 85 (5): 628–42. doi:10.1016/j.ajhg.2009.10.014. PMC 2775832. PMID 19913121.
- Carreño LJ, Pacheco R, Gutierrez MA, Jacobelli S, Kalergis AM (November 2009). "Disease activity in systemic lupus erythematosus is associated with an altered expression of low-affinity Fc gamma receptors and costimulatory molecules on dendritic cells". Immunology. 128 (3): 334–41. doi:10.1111/j.1365-2567.2009.03138.x. PMC 2770681. PMID 20067533.
- Koyasu S (April 2003). "The role of PI3K in immune cells". Nature Immunology. 4 (4): 313–9. doi:10.1038/ni0403-313. PMID 12660731. S2CID 9951653.
- Kim SH, Lee JE, Kim SH, Jee YK, Kim YK, Park HS, et al. (December 2009). "Allelic variants of CD40 and CD40L genes interact to promote antibiotic-induced cutaneous allergic reactions". Clinical and Experimental Allergy. 39 (12): 1852–6. doi:10.1111/j.1365-2222.2009.03336.x. PMID 19735272. S2CID 26024387.
- Liu Y, Liang WB, Gao LB, Pan XM, Chen TY, Wang YY, et al. (November 2010). "CTLA4 and CD86 gene polymorphisms and susceptibility to chronic obstructive pulmonary disease". Human Immunology. 71 (11): 1141–6. doi:10.1016/j.humimm.2010.08.007. PMID 20732370.
- Ma XN, Wang X, Yan YY, Yang L, Zhang DL, Sheng X, et al. (June 2010). "Absence of association between CD86 +1057G/A polymorphism and coronary artery disease". DNA and Cell Biology. 29 (6): 325–8. doi:10.1089/dna.2009.0987. PMID 20230296.
- Ishizaki Y, Yukaya N, Kusuhara K, Kira R, Torisu H, Ihara K, et al. (April 2010). "PD1 as a common candidate susceptibility gene of subacute sclerosing panencephalitis". Human Genetics. 127 (4): 411–9. doi:10.1007/s00439-009-0781-z. PMID 20066438. S2CID 12633836.
- Chang TT, Kuchroo VK, Sharpe AH (2002). "Role of the B7-CD28/CTLA-4 pathway in autoimmune disease". Current Directions in Autoimmunity. 5: 113–30. doi:10.1159/000060550. ISBN 3-8055-7308-1. PMID 11826754.
- Grujic M, Bartholdy C, Remy M, Pinschewer DD, Christensen JP, Thomsen AR (August 2010). "The role of CD80/CD86 in generation and maintenance of functional virus-specific CD8+ T cells in mice infected with lymphocytic choriomeningitis virus". Journal of Immunology. 185 (3): 1730–43. doi:10.4049/jimmunol.0903894. PMID 20601595.
- Quaranta MG, Mattioli B, Giordani L, Viora M (November 2006). "The immunoregulatory effects of HIV-1 Nef on dendritic cells and the pathogenesis of AIDS". FASEB Journal. 20 (13): 2198–208. doi:10.1096/fj.06-6260rev. PMID 17077296. S2CID 3111709.
- Schuurhof A, Bont L, Siezen CL, Hodemaekers H, van Houwelingen HC, Kimman TG, et al. (June 2010). "Interleukin-9 polymorphism in infants with respiratory syncytial virus infection: an opposite effect in boys and girls". Pediatric Pulmonology. 45 (6): 608–13. doi:10.1002/ppul.21229. PMID 20503287. S2CID 24678182.
- Bailey SD, Xie C, Do R, Montpetit A, Diaz R, Mohan V, et al. (October 2010). "Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study". Diabetes Care. 33 (10): 2250–3. doi:10.2337/dc10-0452. PMC 2945168. PMID 20628086.
- Radziewicz H, Ibegbu CC, Hon H, Bédard N, Bruneau J, Workowski KA, et al. (March 2010). "Transient CD86 expression on hepatitis C virus-specific CD8+ T cells in acute infection is linked to sufficient IL-2 signaling". Journal of Immunology. 184 (5): 2410–22. doi:10.4049/jimmunol.0902994. PMC 2924663. PMID 20100932.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.