Eggerthellaceae
| Eggerthellaceae | |
|---|---|
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | Eggerthellales Gupta et al. 2013[1] |
| Family: | Eggerthellaceae Gupta et al. 2013[1] |
| Genera[2] | |
| |
The Eggerthellaceae are a family of Gram-positive, rod- or coccus-shaped Actinomycetota.[1][2] It is the sole family within the order Eggerthellales.[2]
The name Eggerthellaceae is derived from the Latin term Eggerthella, referring to the type genus of the family and the suffix "-ceae," an ending used to denote a family. Together, Eggerthellaceae refers to a family whose nomenclatural type is the genus Eggerthella.[4]
Biochemical characteristics and molecular signatures
Members of this family are mostly anaerobic, non-motile (with the exception of some Gordonibacter and Senegalimassilia species that exhibit motility), asaccharolytic and do not form spores.[4] Eggerthellaceae species are commonly isolated from human and animal faeces and other human sources such as the colon, vagina, oral cavity and blood.[4] Some species have also been isolated from human samples of periodontal or endodontic infections, Crohn's disease and severe blood bacteremia.[4] The G+C content for Eggerthellaceae species is between 43.4 and 66.3 mol%.[4]
Analysis of genome sequences identified 13 conserved signature indels (CSIs) in diverse proteins that are exclusively present in different Eggerthellaceae species. The proteins containing these CSIs are: contaexcinuclease ABC subunit UvrC, DNA polymerase III subunit alpha, ribonuclease III, tRNA (N6-isopentenyl adenosine(37)-C2)-methylthiotransferase (MiaB), peptide chain release factor 2, 16S rRNA (cytosine(1402)-N(4))-methyltransferase, elongation factor Tu, UDP-N-acetylglucosamine 1-carboxyvinyltransferase DNA polymerase III subunit epsilon, UDP-glucose 4-epimerase and DNA-directed RNA polymerase subunit alpha.[1][4] The identified CSIs provide reliable and important means to distinguish members of Eggerthellaceae from all other Coriobacteriia as well as other bacteria.
Phylogeny
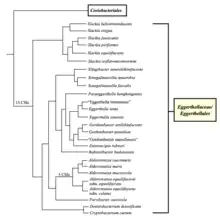
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN).[2] The phylogeny is based on whole-genome analysis.[3][lower-alpha 1] See figure for a detailed phylogenetic tree.
| |||||||||||||||||||||||||||||||||||||||||||
Notes
- ↑ Asaccharobacter, Ellagibacter, Enteroscipio, Paraeggerthella, and Rubneribacter are not included in this phylogenetic tree.
References
- 1 2 3 4 Gupta RS, Chen WJ, Adeolu M, Chai Y. (2013). "Molecular signatures for the class Coriobacteriia and its different clades; Proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov". Int J Syst Evol Microbiol. 63 (Pt 9): 3379–3397. doi:10.1099/ijs.0.048371-0. PMID 23524353.
{{cite journal}}: CS1 maint: uses authors parameter (link) - 1 2 3 4 Euzéby JP, Parte AC. "Eggerthellaceae". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved June 23, 2021.
{{cite web}}: CS1 maint: uses authors parameter (link) - 1 2 Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, Pukall R, Klenk H-P, Goodfellow M, Markus Göker M. (2018). "Genome-Based Taxonomic Classification of the Phylum Actinobacteria". Front. Microbiol. 9: 2007. doi:10.3389/fmicb.2018.02007. PMC 6113628. PMID 30186281.
{{cite journal}}: CS1 maint: uses authors parameter (link) - 1 2 3 4 5 6 Gupta, Radhey S. (2021), Carro, Lorena (ed.), "Eggerthellaceae", Bergey's Manual of Systematics of Archaea and Bacteria, American Cancer Society, pp. 1–11, doi:10.1002/9781118960608.fbm00384, ISBN 978-1-118-96060-8, retrieved 2021-07-05
- ↑ Gupta, Radhey S. (2021), Carro, Lorena (ed.), "Eggerthellaceae", Bergey's Manual of Systematics of Archaea and Bacteria, American Cancer Society, pp. 1–11, doi:10.1002/9781118960608.fbm00384, ISBN 978-1-118-96060-8, retrieved 2021-07-13