Human Microbiome Project
| Human Microbiome Project (HMP) | |
|---|---|
 | |
| Owner | US National Institutes of Health |
| Established | 2007 |
| Disestablished | 2016 |
| Website | hmpdacc |
The Human Microbiome Project (HMP) was a United States National Institutes of Health (NIH) research initiative to improve understanding of the microbiota involved in human health and disease. Launched in 2007,[1] the first phase (HMP1) focused on identifying and characterizing human microbiota. The second phase, known as the Integrative Human Microbiome Project (iHMP) launched in 2014 with the aim of generating resources to characterize the microbiome and elucidating the roles of microbes in health and disease states. The program received $170 million in funding by the NIH Common Fund from 2007 to 2016.[2]
Important components of the HMP were culture-independent methods of microbial community characterization, such as metagenomics (which provides a broad genetic perspective on a single microbial community), as well as extensive whole genome sequencing (which provides a "deep" genetic perspective on certain aspects of a given microbial community, i.e. of individual bacterial species). The latter served as reference genomic sequences — 3000 such sequences of individual bacterial isolates are currently planned — for comparison purposes during subsequent metagenomic analysis. The project also financed deep sequencing of bacterial 16S rRNA sequences amplified by polymerase chain reaction from human subjects.[3]
Introduction
Prior to the HMP launch, it was often reported in popular media and scientific literature that there are about 10 times as many microbial cells and 100 times as many microbial genes in the human body as there are human cells; this figure was based on estimates that the human microbiome includes around 100 trillion bacterial cells and an adult human typically has around 10 trillion human cells.[4] In 2014 the American Academy of Microbiology published a FAQ that emphasized that the number of microbial cells and the number of human cells are both estimates, and noted that recent research had arrived at a new estimate of the number of human cells at around 37 trillion cells, meaning that the ratio of microbial to human cells is probably about 3:1.[4][5] In 2016 another group published a new estimate of ratio as being roughly 1:1 (1.3:1, with "an uncertainty of 25% and a variation of 53% over the population of standard 70 kg males").[6][7]
Despite the staggering number of microbes in and on the human body, little was known about their roles in human health and disease. Many of the organisms that make up the microbiome have not been successfully cultured, identified, or otherwise characterized. Organisms thought to be found in the human microbiome, however, may generally be categorized as bacteria, members of domain Archaea, yeasts, and single-celled eukaryotes as well as various helminth parasites and viruses, the latter including viruses that infect the cellular microbiome organisms (e.g., bacteriophages). The HMP set out to discover and characterize the human microbiome, emphasizing oral, skin, vaginal, gastrointestinal, and respiratory sites.
The HMP will address some of the most inspiring, vexing and fundamental scientific questions today. Importantly, it also has the potential to break down the artificial barriers between medical microbiology and environmental microbiology. It is hoped that the HMP will not only identify new ways to determine health and predisposition to diseases but also define the parameters needed to design, implement and monitor strategies for intentionally manipulating the human microbiota, to optimize its performance in the context of an individual's physiology.[8]
The HMP has been described as "a logical conceptual and experimental extension of the Human Genome Project."[8] In 2007 the HMP was listed on the NIH Roadmap for Medical Research[9] as one of the New Pathways to Discovery. Organized characterization of the human microbiome is also being done internationally under the auspices of the International Human Microbiome Consortium.[10] The Canadian Institutes of Health Research, through the CIHR Institute of Infection and Immunity, is leading the Canadian Microbiome Initiative to develop a coordinated and focused research effort to analyze and characterize the microbes that colonize the human body and their potential alteration during chronic disease states.[11]
Contributing Institutions
The HMP involved participation from many research institutions, including Stanford University, the Broad Institute, Virginia Commonwealth University, Washington University, Northeastern University, MIT, the Baylor College of Medicine, and many others. Contributions included data evaluation, construction of reference sequence data sets, ethical and legal studies, technology development, and more.
Phase One (2007-2014)
The HMP1 included research efforts from many institutions.[12] The HMP1 set the following goals:[13]
- Develop a reference set of microbial genome sequences and to perform preliminary characterization of the human microbiome
- Explore the relationship between disease and changes in the human microbiome
- Develop new technologies and tools for computational analysis
- Establish a resource repository
- Study the ethical, legal, and social implications of human microbiome research
Phase Two (2014-2016)
In 2014, the NIH launched the second phase of the project, known as the Integrative Human Microbiome Project (iHMP). The goal of the iHMP was to produce resources to create a complete characterization of the human microbiome, with a focus on understanding the presence of microbiota in health and disease states.[14] The project mission, as stated by the NIH, was as follows:
The iHMP will create integrated longitudinal datasets of biological properties from both the microbiome and host from three different cohort studies of microbiome-associated conditions using multiple "omics" technologies.[14]
The project encompassed three sub-projects carried out at multiple institutions. Study methods included 16S rRNA gene profiling, whole metagenome shotgun sequencing, whole genome sequencing, metatranscriptomics, metabolomics/lipidomics, and immunoproteomics. The key findings of the iHMP were published in 2019.[15]
Pregnancy & Preterm Birth
The Vaginal Microbiome Consortium team at Virginia Commonwealth University led research on the Pregnancy & Preterm Birth project with a goal of understanding how the microbiome changes during the gestational period and influences the neonatal microbiome. The project was also concerned with the role of the microbiome in the occurrence of preterm births, which, according to the CDC, account for nearly 10% of all births[16] and constitutes the second leading cause of neonatal death.[17] The project received $7.44 million in NIH funding.[18]
Onset of Inflammatory Bowel Disease (IBD)
The Inflammatory Bowel Disease Multi'omics Data (IBDMDB) team was a multi-institution group of researchers focused on understanding how the gut microbiome changes longitudinally in adults and children suffering from IBD. IBD is an inflammatory autoimmune disorder that manifests as either Crohn's disease or ulcerative colitis and affects about one million Americans.[19] Research participants included cohorts from Massachusetts General Hospital, Emory University Hospital/Cincinnati Children's Hospital, and Cedars-Sinai Medical Center.[20]
Onset of Type 2 Diabetes (T2D)
Researchers from Stanford University and the Jackson Laboratory of Genomic Medicine worked together to perform a longitudinal analysis on the biological processes that occur in the microbiome of patients at risk for Type 2 Diabetes. T2D affects nearly 20 million Americans with at least 79 million pre-diabetic patients,[21] and is partially characterized by marked shifts in the microbiome compared to healthy individuals. The project aimed to identify molecules and signaling pathways that play a role in the etiology of the disease.[22]
Achievements
The impact to date of the HMP may be partially assessed by examination of research sponsored by the HMP. Over 650 peer-reviewed publications were listed on the HMP website from June 2009 to the end of 2017, and had been cited over 70,000 times.[23] At this point the website was archived and is no longer updated, although datasets do continue to be available.[24]
Major categories of work funded by HMP included:
- Development of new database systems allowing efficient organization, storage, access, search and annotation of massive amounts of data. These include IMG, the Integrated Microbial Genomes database and comparative analysis system;[25] IMG/M, a related system that integrates metagenome data sets with isolate microbial genomes from the IMG system;[26] CharProtDB, a database of experimentally characterized protein annotations;[27] and the Genomes OnLine Database (GOLD), for monitoring the status of genomic and metagenomic projects worldwide and their associated metadata.[28]
- Development of tools for comparative analysis that facilitate the recognition of common patterns, major themes and trends in complex data sets. These include RAPSearch2, a fast and memory-efficient protein similarity search tool for next-generation sequencing data;[29] Boulder ALignment Editor (ALE), a web-based RNA alignment tool;[30] WebMGA, a customizable web server for fast metagenomic sequence analysis;[31] and DNACLUST, a tool for accurate and efficient clustering of phylogenetic marker genes[32]
- Development of new methods and systems for assembly of massive sequence data sets. No single assembly algorithm addresses all the known problems of assembling short-length sequences,[33] so next-generation assembly programs such as AMOS[34] are modular, offering a wide range of tools for assembly. Novel algorithms have been developed for improving the quality and utility of draft genome sequences.[35]
- Assembly of a catalog of sequenced reference genomes of pure bacterial strains from multiple body sites, against which metagenomic results can be compared. The original goal of 600 genomes has been far surpassed; the current goal is for 3000 genomes to be in this reference catalog, sequenced to at least a high-quality draft stage. As of March 2012, 742 genomes have been cataloged.[36]
- Establishment of the Data Analysis and Coordination Center (DACC),[37] which serves as the central repository for all HMP data.
- Various studies exploring legal and ethical issues associated with whole genome sequencing research.[38][39][40][41]
Developments funded by HMP included:
- New predictive methods for identifying active transcription factor binding sites.[42]
- Identification, on the basis of bioinformatic evidence, of a widely distributed, ribosomally produced electron carrier precursor[43]
- Time-lapse "moving pictures" of the human microbiome.[44]
- Identification of unique adaptations adopted by segmented filamentous bacteria (SFB) in their role as gut commensals.[45] SFB are medically important because they stimulate T helper 17 cells, thought to play a key role in autoimmune disease.
- Identification of factors distinguishing the microbiota of healthy and diseased gut.[46]
- Identification of a hitherto unrecognized dominant role of Verrucomicrobia in soil bacterial communities.[47]
- Identification of factors determining the virulence potential of Gardnerella vaginalis strains in vaginosis.[48]
- Identification of a link between oral microbiota and atherosclerosis.[49]
- Demonstration that pathogenic species of Neisseria involved in meningitis, sepsis, and sexually transmitted disease exchange virulence factors with commensal species.[50]
Milestones
Reference database established
On 13 June 2012, a major milestone of the HMP was announced by the NIH director Francis Collins.[51] The announcement was accompanied with a series of coordinated articles published in Nature[52][53] and several journals including the Public Library of Science (PLoS) on the same day.[54][55][56][57][58][59] By mapping the normal microbial make-up of healthy humans using genome sequencing techniques, the researchers of the HMP have created a reference database and the boundaries of normal microbial variation in humans.[60]
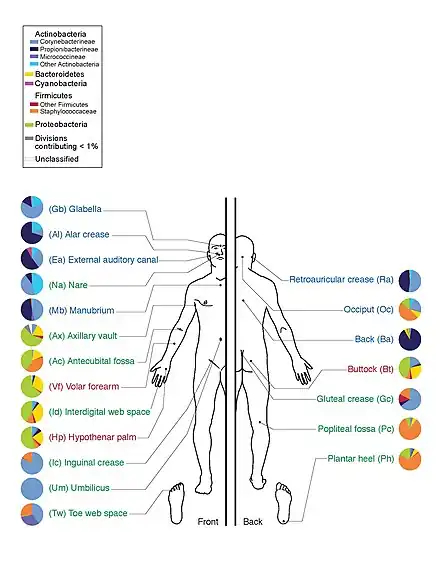
From 242 healthy U.S. volunteers, more than 5,000 samples were collected from tissues from 15 (men) to 18 (women) body sites such as mouth, nose, skin, lower intestine (stool) and vagina. All the DNA, human and microbial, were analyzed with DNA sequencing machines. The microbial genome data were extracted by identifying the bacterial specific ribosomal RNA, 16S rRNA. The researchers calculated that more than 10,000 microbial species occupy the human ecosystem and they have identified 81 – 99% of the genera. In addition to establishing the human microbiome reference database, the HMP project also discovered several "surprises", which include:
- Microbes contribute more genes responsible for human survival than humans' own genes. It is estimated that bacterial protein-coding genes are 360 times more abundant than human genes.
- Microbial metabolic activities; for example, digestion of fats; are not always provided by the same bacterial species. The presence of the activities seems to matter more.
- Components of the human microbiome change over time, affected by a patient disease state and medication. However, the microbiome eventually returns to a state of equilibrium, even though the composition of bacterial types has changed.
Clinical application
Among the first clinical applications utilizing the HMP data, as reported in several PLoS papers, the researchers found a shift to less species diversity in vaginal microbiome of pregnant women in preparation for birth, and high viral DNA load in the nasal microbiome of children with unexplained fevers. Other studies using the HMP data and techniques include role of microbiome in various diseases in the digestive tract, skin, reproductive organs and childhood disorders.[51]
Pharmaceutical application
Pharmaceutical microbiologists have considered the implications of the HMP data in relation to the presence / absence of 'objectionable' microorganisms in non-sterile pharmaceutical products and in relation to the monitoring of microorganisms within the controlled environments in which products are manufactured. The latter also has implications for media selection and disinfectant efficacy studies.[61]
See also
- Environmental microbiology
- Genome project
- Genomics
- Gut flora
- Human microbiome
- Human virome
- Medical microbiology
- Metagenomics
- Microbial ecology
- Microflora
- Multigenomic organism
- Oral microbiology
- Skin flora
- Superorganism
- Vaginal flora
References
- ↑ "Human Microbiome Project: Diversity of Human Microbes Greater Than Previously Predicted". ScienceDaily. Retrieved 8 March 2012.
- ↑ "Human Microbiome Project - Home | NIH Common Fund". commonfund.nih.gov. 28 June 2013. Retrieved 2018-04-15.
- ↑ "Human Microbiome Project". The NIH Common Fund. Retrieved 8 March 2012.
- 1 2 American Academy of Microbiology FAQ: Human Microbiome Archived 2016-12-31 at the Wayback Machine January 2014
- ↑ Judah L. Rosner for Microbe Magazine, Feb 2014. Ten Times More Microbial Cells than Body Cells in Humans?
- ↑ Alison Abbott for Nature News. Jan 8 2016 Scientists bust myth that our bodies have more bacteria than human cells
- ↑ Sender R, Fuchs S, Milo R (January 2016). "Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans". Cell. 164 (3): 337–40. doi:10.1016/j.cell.2016.01.013. PMID 26824647.
- 1 2 Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (October 2007). "The human microbiome project". Nature. 449 (7164): 804–10. Bibcode:2007Natur.449..804T. doi:10.1038/nature06244. PMC 3709439. PMID 17943116.
- ↑ "About the NIH Roadmap". The NIH Common Fund. Archived from the original on 17 February 2013. Retrieved 8 March 2012.
- ↑ "The International Human Microbiome Consortium". Retrieved 8 March 2012.
- ↑ "Canadian Microbiome Initiative". Canadian Institutes of Health Research. 13 August 2009. Retrieved 8 March 2012.
- ↑ "Human Microbiome Project / Funded Research". The NIH Common Fund. Retrieved 8 March 2012.
- ↑ "Human Microbiome Project / Program Initiatives". The NIH Common Fund. Retrieved 8 March 2012.
- 1 2 "NIH Human Microbiome Project - About the Human Microbiome". hmpdacc.org. Retrieved 2018-03-30.
- ↑ NIH Human Microbiome Portfolio Analysis Team (2019). "A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016". Microbiome. 7 (1): 31. doi:10.1186/s40168-019-0620-y. ISSN 2049-2618. PMC 6391833. PMID 30808411.
- ↑ Ferré C, Callaghan W, Olson C, Sharma A, Barfield W (November 2016). "Effects of Maternal Age and Age-Specific Preterm Birth Rates on Overall Preterm Birth Rates - United States, 2007 and 2014". MMWR. Morbidity and Mortality Weekly Report. 65 (43): 1181–1184. doi:10.15585/mmwr.mm6543a1. PMID 27811841.
- ↑ "Infant Mortality | Maternal and Infant Health | Reproductive Health | CDC". www.cdc.gov. 2018-01-02. Retrieved 2018-04-03.
- ↑ Consortium, VCU, Vaginal Microbiome. "Vaginal Microbiome Consortium". vmc.vcu.edu. Retrieved 2018-04-05.
- ↑ "CDC - Epidemiology of the IBD - Inflammatory Bowel Disease". www.cdc.gov. Archived from the original on 2017-02-23. Retrieved 2018-04-15.
- ↑ "Team". ibdmdb.org. Retrieved 2018-04-05.
- ↑ "National Diabetes Statistics Report | Data & Statistics | Diabetes | CDC". www.cdc.gov. 2018-03-09. Retrieved 2018-04-15.
- ↑ "Integrated Personal Omics Profiling | Integrated Personal Omics Profiling | Stanford Medicine". med.stanford.edu. Retrieved 2018-04-05.
- ↑ "Human Microbiome Project - Home | NIH Common Fund". commonfund.nih.gov. 28 June 2013. Retrieved 2019-04-18.
- ↑ "Human Microbiome Project Data Portal". portal.hmpdacc.org. Retrieved 2019-04-18.
- ↑ Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC (January 2012). "IMG: the Integrated Microbial Genomes database and comparative analysis system". Nucleic Acids Research. 40 (Database issue): D115–22. doi:10.1093/nar/gkr1044. PMC 3245086. PMID 22194640.
- ↑ Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Grechkin Y, Ratner A, Jacob B, Pati A, Huntemann M, Liolios K, Pagani I, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC (January 2012). "IMG/M: the integrated metagenome data management and comparative analysis system". Nucleic Acids Research. 40 (Database issue): D123–9. doi:10.1093/nar/gkr975. PMC 3245048. PMID 22086953.
- ↑ Madupu R, Richter A, Dodson RJ, Brinkac L, Harkins D, Durkin S, Shrivastava S, Sutton G, Haft D (January 2012). "CharProtDB: a database of experimentally characterized protein annotations". Nucleic Acids Research. 40 (Database issue): D237–41. doi:10.1093/nar/gkr1133. PMC 3245046. PMID 22140108.
- ↑ Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC (January 2012). "The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata". Nucleic Acids Research. 40 (Database issue): D571–9. doi:10.1093/nar/gkr1100. PMC 3245063. PMID 22135293.
- ↑ Zhao Y, Tang H, Ye Y (January 2012). "RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data". Bioinformatics. 28 (1): 125–6. doi:10.1093/bioinformatics/btr595. PMC 3244761. PMID 22039206.
- ↑ Stombaugh J, Widmann J, McDonald D, Knight R (June 2011). "Boulder ALignment Editor (ALE): a web-based RNA alignment tool". Bioinformatics. 27 (12): 1706–7. doi:10.1093/bioinformatics/btr258. PMC 3106197. PMID 21546392.
- ↑ Wu S, Zhu Z, Fu L, Niu B, Li W (September 2011). "WebMGA: a customizable web server for fast metagenomic sequence analysis". BMC Genomics. 12: 444. doi:10.1186/1471-2164-12-444. PMC 3180703. PMID 21899761.
- ↑ Ghodsi M, Liu B, Pop M (June 2011). "DNACLUST: accurate and efficient clustering of phylogenetic marker genes". BMC Bioinformatics. 12: 271. doi:10.1186/1471-2105-12-271. PMC 3213679. PMID 21718538.
- ↑ Yao G, Ye L, Gao H, Minx P, Warren WC, Weinstock GM (January 2012). "Graph accordance of next-generation sequence assemblies". Bioinformatics. 28 (1): 13–6. doi:10.1093/bioinformatics/btr588. PMC 3244760. PMID 22025481.
- ↑ Treangen TJ, Sommer DD, Angly FE, Koren S, Pop M (March 2011). Next generation sequence assembly with AMOS. Current Protocols in Bioinformatics. Vol. Chapter 11. pp. Unit 11.8. doi:10.1002/0471250953.bi1108s33. ISBN 978-0471250951. PMC 3072823. PMID 21400694.
- ↑ Koren S, Miller JR, Walenz BP, Sutton G (September 2010). "An algorithm for automated closure during assembly". BMC Bioinformatics. 11: 457. doi:10.1186/1471-2105-11-457. PMC 2945939. PMID 20831800.
- ↑ "Human Microbiome Project / Reference Genomes Data". Data Analysis and Coordination Center (DACC) for the National Institutes of Health (NIH). Retrieved 8 March 2012.
- ↑ "Data Analysis and Coordination Center (DACC)". National Institutes of Health (NIH) Common Fund. Retrieved 11 March 2012.
- ↑ Schwab AP, Frank L, Gligorov N (November 2011). "Saying privacy, meaning confidentiality". The American Journal of Bioethics. 11 (11): 44–5. doi:10.1080/15265161.2011.608243. PMID 22047127. S2CID 34313782.
- ↑ Rhodes R, Azzouni J, Baumrin SB, Benkov K, Blaser MJ, Brenner B, Dauben JW, Earle WJ, Frank L, Gligorov N, Goldfarb J, Hirschhorn K, Hirschhorn R, Holzman I, Indyk D, Jabs EW, Lackey DP, Moros DA, Philpott S, Rhodes ME, Richardson LD, Sacks HS, Schwab A, Sperling R, Trusko B, Zweig A (November 2011). "De minimis risk: a proposal for a new category of research risk". The American Journal of Bioethics. 11 (11): 1–7. doi:10.1080/15265161.2011.615588. PMID 22047112. S2CID 205859554.
- ↑ McGuire AL, Lupski JR (May 2010). "Personal genome research : what should the participant be told?". Trends in Genetics. 26 (5): 199–201. doi:10.1016/j.tig.2009.12.007. PMC 2868334. PMID 20381895.
- ↑ Sharp RR, Achkar JP, Brinich MA, Farrell RM (April 2009). "Helping patients make informed choices about probiotics: a need for research". The American Journal of Gastroenterology. 104 (4): 809–13. doi:10.1038/ajg.2008.68. PMC 2746707. PMID 19343022.
- ↑ Cuellar-Partida G, Buske FA, McLeay RC, Whitington T, Noble WS, Bailey TL (January 2012). "Epigenetic priors for identifying active transcription factor binding sites". Bioinformatics. 28 (1): 56–62. doi:10.1093/bioinformatics/btr614. PMC 3244768. PMID 22072382.
- ↑ Haft DH (January 2011). "Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners". BMC Genomics. 12: 21. doi:10.1186/1471-2164-12-21. PMC 3023750. PMID 21223593.
- ↑ Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R (2011). "Moving pictures of the human microbiome". Genome Biology. 12 (5): R50. doi:10.1186/gb-2011-12-5-r50. PMC 3271711. PMID 21624126.
- ↑ Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II (September 2011). "The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment". Cell Host & Microbe. 10 (3): 260–72. doi:10.1016/j.chom.2011.08.005. PMC 3209701. PMID 21925113.
- ↑ Ballal SA, Gallini CA, Segata N, Huttenhower C, Garrett WS (April 2011). "Host and gut microbiota symbiotic factors: lessons from inflammatory bowel disease and successful symbionts". Cellular Microbiology. 13 (4): 508–17. doi:10.1111/j.1462-5822.2011.01572.x. PMID 21314883. S2CID 205529660.
- ↑ Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N (July 2011). "The under-recognized dominance of Verrucomicrobia in soil bacterial communities". Soil Biology & Biochemistry. 43 (7): 1450–1455. doi:10.1016/j.soilbio.2011.03.012. PMC 3260529. PMID 22267877.
- ↑ Yeoman CJ, Yildirim S, Thomas SM, Durkin AS, Torralba M, Sutton G, Buhay CJ, Ding Y, Dugan-Rocha SP, Muzny DM, Qin X, Gibbs RA, Leigh SR, Stumpf R, White BA, Highlander SK, Nelson KE, Wilson BA (August 2010). Li W (ed.). "Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential". PLOS ONE. 5 (8): e12411. Bibcode:2010PLoSO...512411Y. doi:10.1371/journal.pone.0012411. PMC 2928729. PMID 20865041.
- ↑ Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Bäckhed F (March 2011). "Human oral, gut, and plaque microbiota in patients with atherosclerosis". Proceedings of the National Academy of Sciences of the United States of America. 108 Suppl 1 (Supplement_1): 4592–8. Bibcode:2011PNAS..108.4592K. doi:10.1073/pnas.1011383107. PMC 3063583. PMID 20937873.
- ↑ Marri PR, Paniscus M, Weyand NJ, Rendón MA, Calton CM, Hernández DR, Higashi DL, Sodergren E, Weinstock GM, Rounsley SD, So M (July 2010). Ahmed N (ed.). "Genome sequencing reveals widespread virulence gene exchange among human Neisseria species". PLOS ONE. 5 (7): e11835. Bibcode:2010PLoSO...511835M. doi:10.1371/journal.pone.0011835. PMC 2911385. PMID 20676376.
- 1 2 "NIH Human Microbiome Project defines normal bacterial makeup of the body". NIH News. 13 June 2012.
- ↑ Human Microbiome Project Consortium (June 2012). "A framework for human microbiome research". Nature. 486 (7402): 215–21. Bibcode:2012Natur.486..215T. doi:10.1038/nature11209. PMC 3377744. PMID 22699610.
- ↑ Human Microbiome Project Consortium (June 2012). "Structure, function and diversity of the healthy human microbiome". Nature. 486 (7402): 207–14. Bibcode:2012Natur.486..207T. doi:10.1038/nature11234. PMC 3564958. PMID 22699609.
- ↑ Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C (2012). "Microbial co-occurrence relationships in the human microbiome". PLOS Computational Biology. 8 (7): e1002606. Bibcode:2012PLSCB...8E2606F. doi:10.1371/journal.pcbi.1002606. PMC 3395616. PMID 22807668.
- ↑ Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methé B, Schloss PD, Gevers D, Mitreva M, Huttenhower C (2012). "Metabolic reconstruction for metagenomic data and its application to the human microbiome". PLOS Computational Biology. 8 (6): e1002358. Bibcode:2012PLSCB...8E2358A. doi:10.1371/journal.pcbi.1002358. PMC 3374609. PMID 22719234.
- ↑ Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J (June 2012). "Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples". Genome Biology. 13 (6): R42. doi:10.1186/gb-2012-13-6-r42. PMC 3446314. PMID 22698087.
- ↑ Cantarel BL, Lombard V, Henrissat B (2012). "Complex carbohydrate utilization by the healthy human microbiome". PLOS ONE. 7 (6): e28742. Bibcode:2012PLoSO...728742C. doi:10.1371/journal.pone.0028742. PMC 3374616. PMID 22719820.
- ↑ Wu YW, Rho M, Doak TG, Ye Y (August 2012). "Oral spirochetes implicated in dental diseases are widespread in normal human subjects and carry extremely diverse integron gene cassettes". Applied and Environmental Microbiology. 78 (15): 5288–96. doi:10.1128/AEM.00564-12. PMC 3416431. PMID 22635997.
- ↑ "PLOS Collections: Article collections published by the Public Library of Science". collections.plos.org. Retrieved 2018-04-15.
- ↑ Manuscript Summaries
- ↑ Wilder, C, Sandle T, Sutton S (June 2013). "Implications of the Human Microbiome on Pharmaceutical Microbiology". American Pharmaceutical Review.
External links
- Human Microbiome Project
- Data Analysis and Coordination Center
- The CIHR Canadian Microbiome Initiative
- The International Human Microbiome Consortium
- 2006, Lay summary of colon microbiome study (the actual study: Gill et al., 2006)
- Olivia Judson Microbes ‘R’ Us New York Times 22 July 2009
- Gina Kolata Good Health? Thank Your 100 Trillion Bacteria New York Times 13 June 2012