Salivary microbiome
The salivary microbiome consists of the nonpathogenic, commensal bacteria present in the healthy human salivary glands. It differs from the oral microbiome which is located in the oral cavity. Oral microorganisms tend to adhere to teeth.[1] The oral microbiome possesses its own characteristic microorganisms found there. Resident microbes of the mouth adhere to the teeth and gums. "[T]here may be important interactions between the saliva microbiome and other microbiomes in the human body, in particular, that of the intestinal tract."[2]

Characteristics
Unlike the uterine, placental and vaginal microbiomes, the types of organisms in the salivary microbiota remain relatively constant. There is no difference between populations of microbes of based upon gender, age, diet, obesity, alcohol intake, race, or tobacco use.[3] Porphyromonas, Solobacterium, Haemophilus, Corynebacterium, Cellulosimicrobium, Streptococcus and Campylobacter are some of the genera found in the saliva.[4]
Genetic markers and diagnostic testing
"There is high diversity in the salivary microbiome within and between individuals, but little geographic structure. Overall, ∼13.5% of the total variance in the composition of genera is due to differences among individuals, which is remarkably similar to the fraction of the total variance in neutral genetic markers that can be attributed to differences among human populations."[2]
"[E]nvironmental variables revealed a significant association between the genetic distances among locations and the distance of each location from the equator. Further characterization of the enormous diversity revealed here in the human salivary microbiome will aid in elucidating the role it plays in human health and disease, and in the identification of potentially informative species for studies of human population history."[2]
Sixty new genera have been identified from the salivary glands. A total of 101 different genera were identified in the salivary glands. Out of these, 39 genera are not found in the oral microbiome. It is not known whether the resident species remain constant or change.[2]
Though the association between the salivary microbiome is similar to that of the oral microbiome, there also exists an association the salivary microbiome and the gut microbiome. Saliva sampling may be a non-invasive way to detect changes in the gut microbiome and changes in systemic disease. The association between the salivary microbiome those with Polycistic Ovarian Syndrome has been characterized: "saliva microbiome profiles correlate with those in the stool, despite the fact that the bacterial communities in the two locations differ greatly. Therefore, saliva may be a useful alternative to stool as an indicator of bacterial dysbiosis in systemic disease."[5]
The sugar concentration in salivary secretions can vary. Blood sugar levels are reflected in salivary gland secretions. High salivary glucose (HSG) levels are a glucose concentration ≥ 1.0 mg/d, n = 175) and those with low salivary glucose (LSG) levels are < 0.1 mg/dL n = 2,537). Salivary gland secretions containing high levels of sugar change the oral microbiome and contributes to an environment that is conductive to the formation of dental caries and gingivitis.[6]
Salivary glands
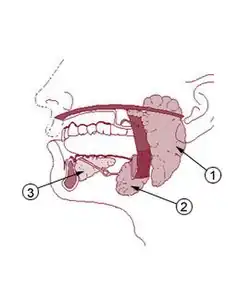
Organisms of the salivary microbiome reside in the three major salivary glands: parotid, submandibular, and sublingual. These glands secrete electrolytes, proteins, genetic material, polysaccharides, and other molecules. Most of these substances enter the salivary gland acinus and duct system from surrounding capillaries via the intervening tissue fluid, although some substances are produced within the glands themselves. The level of each salivary component varies considerably depending on the health status of the individual and the presence of pathogenic and commensal organisms.
References
- ↑ Schwiertz, Andreas (2016). Microbiota of the human body : implications in health and disease. Switzerland: Springer. p. 45. ISBN 978-3-319-31248-4.
- 1 2 3 4 Nasidze, Ivan; Li, Jing; Quinque, Dominique; Tang, Kun; Stoneking, Mark (1 April 2009). "Global diversity in the human salivary microbiome". Genome Research. 19 (4): 636–643. doi:10.1101/gr.084616.108. PMC 2665782. PMID 19251737. Retrieved 6 May 2017 – via genome.cshlp.org.
- ↑ Lindheim, Lisa; Bashir, Mina; Münzker, Julia; Trummer, Christian; Zachhuber, Verena; Pieber, Thomas R.; Gorkiewicz, Gregor; Obermayer-Pietsch, Barbara (25 August 2016). "The Salivary Microbiome in Polycystic Ovary Syndrome (PCOS) and Its Association with Disease-Related Parameters: A Pilot Study". Frontiers in Microbiology. 7 Central: 1270. doi:10.3389/fmicb.2016.01270. PMC 4996828. PMID 27610099.
- ↑ Wang, Kun; Lu, Wenxin; Tu, Qichao; Ge, Yichen; He, Jinzhi; Zhou, Yu; Gou, Yaping; Nostrand, Joy D Van; Qin, Yujia; Li, Jiyao; Zhou, Jizhong; Li, Yan; Xiao, Liying; Zhou, Xuedong (10 March 2016). "Preliminary analysis of salivary microbiome and their potential roles in oral lichen planus". Scientific Reports. 6 (1): 22943. Bibcode:2016NatSR...622943W. doi:10.1038/srep22943. PMC 4785528. PMID 26961389.
- ↑ Lindheim, Lisa; Bashir, Mina; Münzker, Julia; Trummer, Christian; Zachhuber, Verena; Pieber, Thomas R.; Gorkiewicz, Gregor; Obermayer-Pietsch, Barbara (1 January 2016). "The Salivary Microbiome in Polycystic Ovary Syndrome (PCOS) and Its Association with Disease-Related Parameters: A Pilot Study". Frontiers in Microbiology. 7: 1270. doi:10.3389/fmicb.2016.01270. PMC 4996828. PMID 27610099.
- ↑ Nascimento, Marcelle; Goodson, J. Max; Hartman, Mor-Li; Shi, Ping; Hasturk, Hatice; Yaskell, Tina; Vargas, Jorel; Song, Xiaoqing; Cugini, Maryann; Barake, Roula; Alsmadi, Osama; Al-Mutawa, Sabiha; Ariga, Jitendra; Soparkar, Pramod; Behbehani, Jawad; Behbehani, Kazem (2017). "The salivary microbiome is altered in the presence of a high salivary glucose concentration". PLOS ONE. 12 (3): e0170437. Bibcode:2017PLoSO..1270437G. doi:10.1371/journal.pone.0170437. ISSN 1932-6203. PMC 5331956. PMID 28249034.