Plant microbiome
| Part of a series on |
| Microbiomes |
|---|
 |
The plant microbiome, also known as the phytomicrobiome, plays roles in plant health and productivity and has received significant attention in recent years.[1][2] The microbiome has been defined as "a characteristic microbial community occupying a reasonably well-defined habitat which has distinct physio-chemical properties. The term thus not only refers to the microorganisms involved but also encompasses their theatre of activity".[3][4]
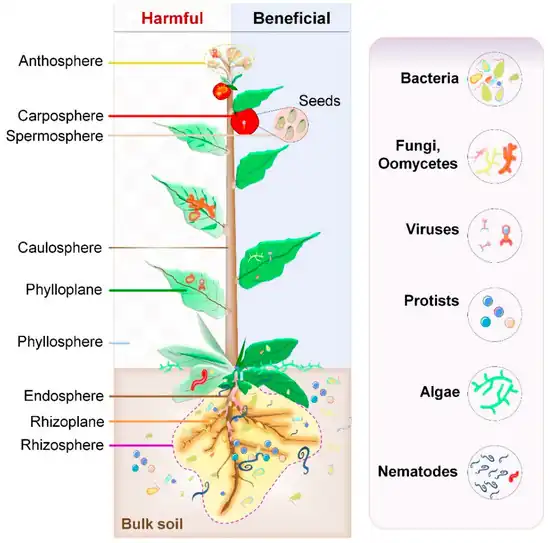
Plants live in association with diverse microbial consortia. These microbes, referred to as the plant's microbiota, live both inside (the endosphere) and outside (the episphere) of plant tissues, and play important roles in the ecology and physiology of plants.[5] "The core plant microbiome is thought to comprise keystone microbial taxa that are important for plant fitness and established through evolutionary mechanisms of selection and enrichment of microbial taxa containing essential functions genes for the fitness of the plant holobiont."[6]
Plant microbiomes are shaped by both factors related to the plant itself, such as genotype, organ, species and health status, as well as factors related to the plant's environment, such as management, land use and climate.[7] The health status of a plant has been reported in some studies to be reflected by or linked to its microbiome.[8][1][9][2]
Overview
The study of the association of plants with microorganisms precedes that of the animal and human microbiomes, notably the roles of microbes in nitrogen and phosphorus uptake. The most notable examples are plant root-arbuscular mycorrhizal (AM) and legume-rhizobial symbioses, both of which greatly influence the ability of roots to uptake various nutrients from the soil. Some of these microbes cannot survive in the absence of the plant host (obligate symbionts include viruses and some bacteria and fungi), which provides space, oxygen, proteins, and carbohydrates to the microorganisms. The association of AM fungi with plants has been known since 1842, and over 80% of land plants are found associated with them.[11] It is thought AM fungi helped in the domestication of plants.[5]
Traditionally, plant-microbe interaction studies have been confined to culturable microbes. The numerous microbes that couldn't be cultured have remained uninvestigated, so knowledge of their roles is largely unknown.[5] The possibilities of unraveling the types and outcomes of these plant-microbe interactions has generated considerable interest among ecologists, evolutionary biologists, plant biologists, and agronomists.[12][13][1] Recent developments in multiomics and the establishment of large collections of microorganisms have dramatically increased knowledge of the plant microbiome composition and diversity. The sequencing of marker genes of entire microbial communities, referred to as metagenomics, sheds light on the phylogenetic diversity of the microbiomes of plants. It also adds to the knowledge of the major biotic and abiotic factors responsible for shaping plant microbiome community assemblages.[13][5]
The composition of microbial communities associated with different plant species is correlated with the phylogenetic distance between the plant species, that is, closely related plant species tend to have more alike microbial communities than distant species.[14] The focus of plant microbiome studies has been directed at model plants, such as Arabidopsis thaliana, as well as important economic crop species including barley (Hordeum vulgare), corn (Zea mays), rice (Oryza sativa), soybean (Glycine max), wheat (Triticum aestivum), whereas less attention has been given to fruit crops and tree species.[15][2]
Plant microbiota
Cyanobacteria are an example of a microorganism which widely interacts in a symbiotic manner with land plants.[16][17][18][19] Cyanobacteria can enter the plant through the stomata and colonise the intercellular space, forming loops and intracellular coils.[20] Anabaena spp. colonize the roots of wheat and cotton plants.[21][22][23] Calothrix sp. has also been found on the root system of wheat.[22][23] Monocots, such as wheat and rice, have been colonised by Nostoc spp.,[24][25][26][27] In 1991, Ganther and others isolated diverse heterocystous nitrogen-fixing cyanobacteria, including Nostoc, Anabaena and Cylindrospermum, from plant root and soil. Assessment of wheat seedling roots revealed two types of association patterns: loose colonization of root hair by Anabaena and tight colonization of the root surface within a restricted zone by Nostoc.[24][28]

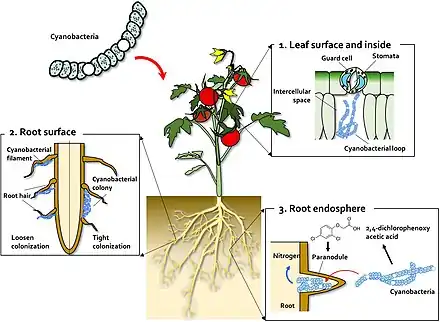
(2) On the root surface, cyanobacteria exhibit two types of colonization pattern; in the root hair, filaments of Anabaena and Nostoc species form loose colonies, and in the restricted zone on the root surface, specific Nostoc species form cyanobacterial colonies.
(3) Co-inoculation with 2,4-D and Nostoc spp. increases para-nodule formation and nitrogen fixation. A large number of Nostoc spp. isolates colonize the root endosphere and form para-nodules.[28]
Rhizosphere microbiome
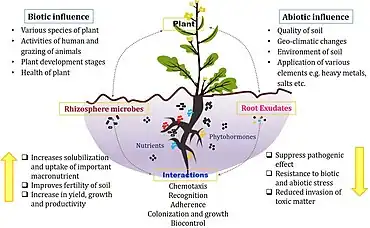
The rhizosphere comprises the 1–10 mm zone of soil immediately surrounding the roots that is under the influence of the plant through its deposition of root exudates, mucilage and dead plant cells.[31] A diverse array of organisms specialize in living in the rhizosphere, including bacteria, fungi, oomycetes, nematodes, algae, protozoa, viruses, and archaea.[32]
"Experimental evidence underlines the importance of the root microbiome in plant health and it is becoming increasingly clear that the plant is able to control the composition of its microbiome. It stands to reason that those plants that manage their microbiome in a way that is beneficial to their reproductive success will be favored during evolutionary selection. It appears that such selective pressure has brought about many specific interactions between plants and microbes, and evidence is accumulating that plants call for microbial help in time of need."
– Berendsen et al, 2012 [33]
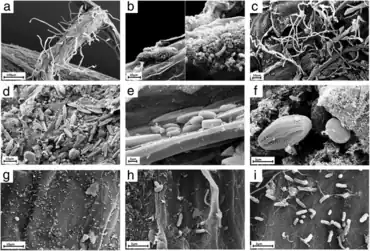
on the roots of Arabidopsis thaliana
a) Overview of an A. thaliana root (primary root) with numerous root hairs. b) Biofilm-forming bacteria. c) Fungal or oomycete hyphae surrounding the root surface. d) Primary root densely covered by spores and protists. e, f) Protists, most likely belonging to the Bacillariophyceae class. g) Bacteria and bacterial filaments. h, i) Different bacterial individuals showing great varieties of shapes and morphological features.[34]
Mycorrhizal fungi are abundant members of the rhizosphere community, and have been found in over 200,000 plant species, and are estimated to associate with over 80% of all plants.[35] Mycorrhizae–root associations play profound roles in land ecosystems by regulating nutrient and carbon cycles. Mycorrhizae are integral to plant health because they provide up to 80% of the nitrogen and phosphorus requirements. In return, the fungi obtain carbohydrates and lipids from host plants.[36] Recent studies of arbuscular mycorrhizal fungi using sequencing technologies show greater between-species and within-species diversity than previously known.[37][5]
The most frequently studied beneficial rhizosphere organisms are mycorrhizae, rhizobium bacteria, plant-growth promoting rhizobacteria (PGPR), and biocontrol microbes. It has been projected that one gram of soil could contain more than one million distinct bacterial genomes,[38] and over 50,000 OTUs (operational taxonomic units) have been found within the potato rhizosphere.[39] Among the prokaryotes in the rhizosphere, the most frequent bacteria are within the Acidobacteria, Proteobacteria, Planctomycetes, Actinobacteria, Bacteroidetes, and Firmicutes.[40][41] In some studies, no significant differences were reported in the microbial community composition between the bulk soil (soil not attached to the plant root) and rhizosphere soil.[42][43] Certain bacterial groups (e. g. Actinobacteria, Xanthomonadaceae) are less abundant in the rhizosphere than in nearby bulk soil .[40][5]
Phyllosphere microbiome
The aerial surface of a plant (stem, leaf, flower, fruit) is called the phyllosphere and is considered comparatively nutrient poor when compared to the rhizosphere and endosphere. The environment in the phyllosphere is more dynamic than the rhizosphere and endosphere environments. Microbial colonizers are subjected to diurnal and seasonal fluctuations of heat, moisture, and radiation. In addition, these environmental elements affect plant physiology (such as photosynthesis, respiration, water uptake etc.) and indirectly influence microbiome composition.[5] Rain and wind also cause temporal variation to the phyllosphere microbiome.[45]
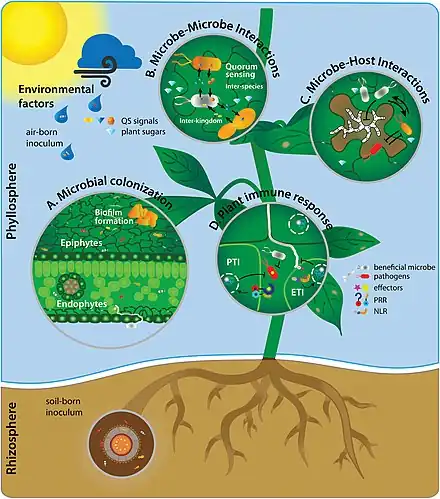
Interactions between plants and their associated microorganisms in many of these microbiomes can play pivotal roles in host plant health, function, and evolution.[46] The leaf surface, or phyllosphere, harbours a microbiome comprising diverse communities of bacteria, fungi, algae, archaea, and viruses.[47][48] Interactions between the host plant and phyllosphere bacteria have the potential to drive various aspects of host plant physiology.[49][50][51] However, as of 2020 knowledge of these bacterial associations in the phyllosphere remains relatively modest, and there is a need to advance fundamental knowledge of phyllosphere microbiome dynamics.[52][53]
Overall, there remains high species richness in phyllosphere communities. Fungal communities are highly variable in the phyllosphere of temperate regions and are more diverse than in tropical regions.[54] There can be up to 107 microbes per square centimetre present on the leaf surfaces of plants, and the bacterial population of the phyllosphere on a global scale is estimated to be 1026 cells.[55] The population size of the fungal phyllosphere is likely to be smaller.[56]
Phyllosphere microbes from different plants appear to be somewhat similar at high levels of taxa, but at the lower levels taxa there remain significant differences. This indicates microorganisms may need finely tuned metabolic adjustment to survive in phyllosphere environment.[54] Proteobacteria seems to be the dominant colonizers, with Bacteroidetes and Actinobacteria also predominant in phyllospheres.[57] Although there are similarities between the rhizosphere and soil microbial communities, very little similarity has been found between phyllosphere communities and microorganisms floating in open air (aeroplankton).[58][5]
The assembly of the phyllosphere microbiome, which can be strictly defined as epiphytic bacterial communities on the leaf surface, can be shaped by the microbial communities present in the surrounding environment (i.e., stochastic colonisation) and the host plant (i.e., biotic selection).[47][59][53] However, although the leaf surface is generally considered a discrete microbial habitat,[60][61] there is no consensus on the dominant driver of community assembly across phyllosphere microbiomes. For example, host-specific bacterial communities have been reported in the phyllosphere of co-occurring plant species, suggesting a dominant role of host selection.[61][62][63][53]
Conversely, microbiomes of the surrounding environment have also been reported to be the primary determinant of phyllosphere community composition.[60][64][65][66] As a result, the processes that drive phyllosphere community assembly are not well understood but unlikely to be universal across plant species. However, the existing evidence does indicate that phyllosphere microbiomes exhibiting host-specific associations are more likely to interact with the host than those primarily recruited from the surrounding environment.[49][67][68][69][53]
The search for a core microbiome in host-associated microbial communities is a useful first step in trying to understand the interactions that may be occurring between a host and its microbiome.[70][71] The prevailing core microbiome concept is built on the notion that the persistence of a taxon across the spatiotemporal boundaries of an ecological niche is directly reflective of its functional importance within the niche it occupies; it therefore provides a framework for identifying functionally critical microorganisms that consistently associate with a host species.[70][72][73][53]
Divergent definitions of “core microbiome” have arisen across scientific literature with researchers variably identifying “core taxa” as those persistent across distinct host microhabitats [74][75] and even different species.[63][67] Given the functional divergence of microorganisms across different host species [63] and microhabitats,[76] defining core taxa sensu stricto as those persistent across broad geographic distances within tissue- and species-specific host microbiomes, represents the most biologically and ecologically appropriate application of this conceptual framework.[77][53] Tissue- and species-specific core microbiomes across host populations separated by broad geographical distances have not been widely reported for the phyllosphere using the stringent definition established by Ruinen.[50][53]
Example: The mānuka phyllosphere
.jpg.webp)
.png.webp)
The flowering tea tree commonly known as mānuka is indigenous to New Zealand.[78] Mānuka honey, produced from the nectar of mānuka flowers, is known for its non-peroxide antibacterial properties.[79][80] These non-peroxide antibacterial properties have been principally linked to the accumulation of the three-carbon sugar dihydroxyacetone (DHA) in the nectar of the mānuka flower, which undergoes a chemical conversion to methylglyoxal (MGO) in mature honey.[81][82][83] However, the concentration of DHA in the nectar of mānuka flowers is notoriously variable, and the antimicrobial efficacy of mānuka honey consequently varies from region to region and from year to year.[84][85][86] Despite extensive research efforts, no reliable correlation has been identified between DHA production and climatic,[87] edaphic,[88] or host genetic factors.[89][53]
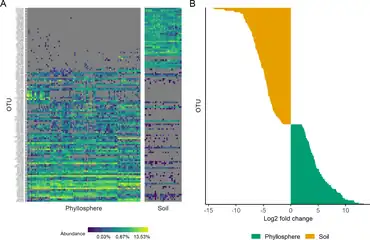
(B) The chart on the right shows how OTUs in phyllosphere and associated soil communities differed in relative abundances.[53]
Microorganisms have been studied in the mānuka rhizosphere and endosphere.[90][91][92] Earlier studies primarily focussed on fungi, and a 2016 study provided the first investigation of endophytic bacterial communities from three geographically and environmentally distinct mānuka populations using fingerprinting techniques and revealed tissue-specific core endomicrobiomes.[93][53] A 2020 study identified a habitat-specific and relatively abundant core microbiome in the mānuka phyllosphere, which was persistent across all samples. In contrast, non-core phyllosphere microorganisms exhibited significant variation across individual host trees and populations that was strongly driven by environmental and spatial factors. The results demonstrated the existence of a dominant and ubiquitous core microbiome in the phyllosphere of mānuka.[53]
Endosphere microbiome
Some microorganisms, such as endophytes, penetrate and occupy the plant internal tissues, forming the endospheric microbiome. The arbuscular mycorrhizal and other endophytic fungi are the dominant colonizers of the endosphere.[94] Bacteria, and to some degree archaea, are important members of endosphere communities. Some of these endophytic microbes interact with their host and provide obvious benefits to plants.[40][95][96] Unlike the rhizosphere and the rhizoplane, the endospheres harbor highly specific microbial communities. The root endophytic community can be very distinct from that of the adjacent soil community. In general, diversity of the endophytic community is lower than the diversity of the microbial community outside the plant.[43] The identity and diversity of the endophytic microbiome of above-and below-ground tissues may also differ within the plant.[97][94][5]
The seed microbiome
Individual seeds possessed high microbial diversity, that was higher in the embryo than in the pericarp. Plant seeds can serve as natural vectors for vertical transmission of beneficial endophytes that confer disease resistance.[98] Evidence showing the transmission of microorganisms from seeds to the developing seedling, was found to occur in experimental and natural conditions.[99] It also seems that the transmission to new plant happens through specific mechanisms where certain microorganisms migrate from seed to plant leaves and other to plant roots.[100]
Plant holobiont
Since the colonization of land by ancestral plant lineages 450 million years ago, plants and their associated microbes have been interacting with each other, forming an assemblage of species that is often referred to as a holobiont. Selective pressure acting on holobiont components has likely shaped plant-associated microbial communities and selected for host-adapted microorganisms that impact plant fitness. However, the high microbial densities detected on plant tissues, together with the fast generation time of microbes and their more ancient origin compared to their host, suggest that microbe-microbe interactions are also important selective forces sculpting complex microbial assemblages in the phyllosphere, rhizosphere, and plant endosphere compartments.[101]
See also
- Biomass partitioning
References
- 1 2 3 Turner, Thomas R.; James, Euan K.; Poole, Philip S. (2013). "The plant microbiome". Genome Biology. 14 (6): 209. doi:10.1186/gb-2013-14-6-209. PMC 3706808. PMID 23805896.
- 1 2 3 Purahong, Witoon; Orrù, Luigi; Donati, Irene; Perpetuini, Giorgia; Cellini, Antonio; Lamontanara, Antonella; Michelotti, Vania; Tacconi, Gianni; Spinelli, Francesco (2018). "Plant Microbiome and Its Link to Plant Health: Host Species, Organs and Pseudomonas syringae pv. Actinidiae Infection Shaping Bacterial Phyllosphere Communities of Kiwifruit Plants". Frontiers in Plant Science. 9: 1563. doi:10.3389/fpls.2018.01563. PMC 6234494. PMID 30464766..
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - ↑ Whipps J., Lewis K. and Cooke R. (1988) "Mycoparasitism and plant disease control". In: Burge M (Ed.) Fungi in Biological Control Systems, Manchester University Press, pages 161–187. ISBN 9780719019791.
- ↑ Berg, Gabriele; Daria Rybakova, Doreen Fischer, Tomislav Cernava, Marie-Christine Champomier Vergès, Trevor Charles, Xiaoyulong Chen, Luca Cocolin, Kellye Eversole, Gema Herrero Corral, Maria Kazou, Linda Kinkel, Lene Lange, Nelson Lima, Alexander Loy, James A. Macklin, Emmanuelle Maguin, Tim Mauchline, Ryan McClure, Birgit Mitter, Matthew Ryan, Inga Sarand, Hauke Smidt, Bettina Schelkle, Hugo Roume, G. Seghal Kiran, Joseph Selvin, Rafael Soares Correa de Souza, Leo van Overbeek, Brajesh K. Singh, Michael Wagner, Aaron Walsh, Angela Sessitsch and Michael Schloter (2020) "Microbiome definition re-visited: old concepts and new challenges". Microbiome, 8(103): 1–22. doi:10.1186/s40168-020-00875-0.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - 1 2 3 4 5 6 7 8 9 10 Dastogeer, K.M., Tumpa, F.H., Sultana, A., Akter, M.A. and Chakraborty, A. (2020) "Plant microbiome–an account of the factors that shape community composition and diversity". Current Plant Biology: 100161. doi:10.1016/j.cpb.2020.100161.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - ↑ Compant, S., Samad, A., Faist, H. and Sessitsch, A. (2019) "A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application". Journal of advanced research, 19: 29_37.doi:10.1016/j.jare.2019.03.004.
- ↑ Bringel, Franã§Oise; Couã©e, Ivan (2015). "Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics". Frontiers in Microbiology. 06: 486. doi:10.3389/fmicb.2015.00486. PMC 4440916. PMID 26052316.
- ↑ Berendsen, Roeland L.; Pieterse, Corné M.J.; Bakker, Peter A.H.M. (2012). "The rhizosphere microbiome and plant health". Trends in Plant Science. 17 (8): 478–486. doi:10.1016/j.tplants.2012.04.001. hdl:1874/255269. PMID 22564542.
- ↑ Berg, Gabriele; Grube, M.; Schloter, M.; Smalla, K. (2014). "The plant microbiome and its importance for plant and human health". Frontiers in Microbiology. 5: 491. doi:10.3389/fmicb.2014.00491. PMC 4166366. PMID 25278934.
- ↑ Shelake, R.M., Pramanik, D. and Kim, J.Y. (2019) "Exploration of plant-microbe interactions for sustainable agriculture in CRISPR era". Microorganisms, 7(8): 269. doi:10.3390/microorganisms7080269.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - ↑ Koide, Roger T.; Mosse, Barbara (2004). "A history of research on arbuscular mycorrhiza". Mycorrhiza. 14 (3): 145–163. doi:10.1007/s00572-004-0307-4. PMID 15088135. S2CID 1809402.
- ↑ Berendsen, Roeland L.; Pieterse, Corné M.J.; Bakker, Peter A.H.M. (2012). "The rhizosphere microbiome and plant health". Trends in Plant Science. 17 (8): 478–486. doi:10.1016/j.tplants.2012.04.001. hdl:1874/255269. PMID 22564542.
- 1 2 Bulgarelli, Davide; Schlaeppi, Klaus; Spaepen, Stijn; Van Themaat, Emiel Ver Loren; Schulze-Lefert, Paul (2013). "Structure and Functions of the Bacterial Microbiota of Plants". Annual Review of Plant Biology. 64: 807–838. doi:10.1146/annurev-arplant-050312-120106. PMID 23373698.
- ↑ Abdelfattah, Ahmed; Tack, Ayco J. M.; Wasserman, Birgit; Liu, Jia; Berg, Gabriele; Norelli, John; Droby, Samir; Wisniewski, Michael (2021). "Evidence for host–microbiome co-evolution in apple". New Phytologist. n/a (n/a). doi:10.1111/nph.17820. ISSN 1469-8137. PMID 34823272. S2CID 244661193.
- ↑ Busby, Posy E.; Soman, Chinmay; Wagner, Maggie R.; Friesen, Maren L.; Kremer, James; Bennett, Alison; Morsy, Mustafa; Eisen, Jonathan A.; Leach, Jan E.; Dangl, Jeffery L. (2017). "Research priorities for harnessing plant microbiomes in sustainable agriculture". PLOS Biology. 15 (3): e2001793. doi:10.1371/journal.pbio.2001793. PMC 5370116. PMID 28350798. S2CID 6434145.
- ↑ Gantar, M.; Elhai, J. (1999). "Colonization of wheatpara-nodules by the N2-fixing cyanobacterium Nostocsp. Strain 2S9B". New Phytologist. 141 (3): 373–379. doi:10.1046/j.1469-8137.1999.00352.x.
- ↑ Gantar, M. (2000). "Mechanical damage of roots provides enhanced colonization of the wheat endorhizosphere by the dinitrogen-fixing cyanobacterium Nostoc sp. Strain 2S9B". Biology and Fertility of Soils. 32 (3): 250–255. doi:10.1007/s003740000243. S2CID 7590731.
- ↑ Treves, Haim; Raanan, Hagai; Kedem, Isaac; Murik, Omer; Keren, Nir; Zer, Hagit; Berkowicz, Simon M.; Giordano, Mario; Norici, Alessandra; Shotland, Yoram; Ohad, Itzhak; Kaplan, Aaron (2016). "The mechanisms whereby the green alga Chlorella ohadii , isolated from desert soil crust, exhibits unparalleled photodamage resistance". New Phytologist. 210 (4): 1229–1243. doi:10.1111/nph.13870. PMID 26853530.
- ↑ Zhu, Huan; Li, Shuyin; Hu, Zhengyu; Liu, Guoxiang (2018). "Molecular characterization of eukaryotic algal communities in the tropical phyllosphere based on real-time sequencing of the 18S rDNA gene". BMC Plant Biology. 18 (1): 365. doi:10.1186/s12870-018-1588-7. PMC 6299628. PMID 30563464.
- ↑ Krings, Michael; Hass, Hagen; Kerp, Hans; Taylor, Thomas N.; Agerer, Reinhard; Dotzler, Nora (2009). "Endophytic cyanobacteria in a 400-million-yr-old land plant: A scenario for the origin of a symbiosis?". Review of Palaeobotany and Palynology. 153 (1–2): 62–69. doi:10.1016/j.revpalbo.2008.06.006.
- ↑ Karthikeyan, N.; Prasanna, R.; Sood, A.; Jaiswal, P.; Nayak, S.; Kaushik, B. D. (2009). "Physiological characterization and electron microscopic investigation of cyanobacteria associated with wheat rhizosphere". Folia Microbiologica. 54 (1): 43–51. doi:10.1007/s12223-009-0007-8. PMID 19330544. S2CID 23420342.
- 1 2 Babu, Santosh; Prasanna, Radha; Bidyarani, Ngangom; Singh, Rajendra (2015). "Analysing the colonisation of inoculated cyanobacteria in wheat plants using biochemical and molecular tools". Journal of Applied Phycology. 27: 327–338. doi:10.1007/s10811-014-0322-6. S2CID 17353123.
- 1 2 Bidyarani, Ngangom; Prasanna, Radha; Chawla, Gautam; Babu, Santosh; Singh, Rajendra (2015). "Deciphering the factors associated with the colonization of rice plants by cyanobacteria". Journal of Basic Microbiology. 55 (4): 407–419. doi:10.1002/jobm.201400591. PMID 25515189. S2CID 5401526.
- 1 2 Gantar, M.; Kerby, N. W.; Rowell, P. (1991). "Colonization of wheat (Triticum vulgare L.) by N2-fixing cyanobacteria: II. An ultrastructural study". New Phytologist. 118 (3): 485–492. doi:10.1111/j.1469-8137.1991.tb00031.x.
- ↑ Ahmed, Mehboob; Stal, Lucas J.; Hasnain, Shahida (2010). "Association of non-heterocystous cyanobacteria with crop plants". Plant and Soil. 336 (1–2): 363–375. doi:10.1007/s11104-010-0488-x. S2CID 21309970.
- ↑ Hussain, Anwar; Hamayun, Muhammad; Shah, Syed Tariq (2013). "Root Colonization and Phytostimulation by Phytohormones Producing Entophytic Nostoc sp. AH-12". Current Microbiology. 67 (5): 624–630. doi:10.1007/s00284-013-0408-4. PMID 23794014. S2CID 14704537.
- ↑ Hussain, Anwar; Shah, Syed T.; Rahman, Hazir; Irshad, Muhammad; Iqbal, Amjad (2015). "Effect of IAA on in vitro growth and colonization of Nostoc in plant roots". Frontiers in Plant Science. 6: 46. doi:10.3389/fpls.2015.00046. PMC 4318279. PMID 25699072.
- 1 2 3 Lee, Sang-Moo; Ryu, Choong-Min (4 February 2021). "Algae as New Kids in the Beneficial Plant Microbiome". Frontiers in Plant Science. Frontiers Media SA. 12: 599742. doi:10.3389/fpls.2021.599742. ISSN 1664-462X. PMC 7889962. PMID 33613596.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - 1 2 Chaudhry, Vasvi; Runge, Paul; Sengupta, Priyamedha; Doehlemann, Gunther; Parker, Jane E; Kemen, Eric (10 September 2020). Kopriva, Stanislav (ed.). "Shaping the leaf microbiota: plant–microbe–microbe interactions". Journal of Experimental Botany. Oxford University Press (OUP). 72 (1): 36–56. doi:10.1093/jxb/eraa417. ISSN 0022-0957. PMC 8210630. PMID 32910810.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - ↑ Vishwakarma, Kanchan; Kumar, Nitin; Shandilya, Chitrakshi; Mohapatra, Swati; Bhayana, Sahil; Varma, Ajit (2020). "Revisiting Plant–Microbe Interactions and Microbial Consortia Application for Enhancing Sustainable Agriculture: A Review". Frontiers in Microbiology. 11: 560406. doi:10.3389/fmicb.2020.560406. PMC 7779480. PMID 33408698.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - ↑ Hinsinger, Philippe; Bengough, A. Glyn; Vetterlein, Doris; Young, Iain M. (2009). "Rhizosphere: Biophysics, biogeochemistry and ecological relevance". Plant and Soil. 321 (1–2): 117–152. doi:10.1007/s11104-008-9885-9. S2CID 8997382.
- ↑ Bonkowski, Michael; Villenave, Cécile; Griffiths, Bryan (2009). "Rhizosphere fauna: The functional and structural diversity of intimate interactions of soil fauna with plant roots". Plant and Soil. 321 (1–2): 213–233. doi:10.1007/s11104-009-0013-2. S2CID 35701713.
- ↑ Berendsen, R.L., Pieterse, C.M. and Bakker, P.A. (2012) "The rhizosphere microbiome and plant health". Trends in plant science, 17(8): 478–486. doi:10.1016/j.tplants.2012.04.001.
- ↑ Hassani, M.A., Durán, P. and Hacquard, S. (2018) "Microbial interactions within the plant holobiont". Microbiome, 6(1): 58. doi:10.1186/s40168-018-0445-0.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - ↑ Van Der Heijden, Marcel G. A.; Martin, Francis M.; Selosse, Marc-André; Sanders, Ian R. (2015). "Mycorrhizal ecology and evolution: The past, the present, and the future". New Phytologist. 205 (4): 1406–1423. doi:10.1111/nph.13288. PMID 25639293.
- ↑ Rich, Mélanie K.; Nouri, Eva; Courty, Pierre-Emmanuel; Reinhardt, Didier (2017). "Diet of Arbuscular Mycorrhizal Fungi: Bread and Butter?" (PDF). Trends in Plant Science. 22 (8): 652–660. doi:10.1016/j.tplants.2017.05.008. PMID 28622919.
- ↑ Lee, Eun-Hwa; Eo, Ju-Kyeong; Ka, Kang-Hyeon; Eom, Ahn-Heum (2013). "Diversity of Arbuscular Mycorrhizal Fungi and Their Roles in Ecosystems". Mycobiology. 41 (3): 121–125. doi:10.5941/MYCO.2013.41.3.121. PMC 3817225. PMID 24198665.
- ↑ Gans, J.; Wolinsky, M.; Dunbar, J. (2005). "Computational Improvements Reveal Great Bacterial Diversity and High Metal Toxicity in Soil". Science. 309 (5739): 1387–1390. Bibcode:2005Sci...309.1387G. doi:10.1126/science.1112665. PMID 16123304. S2CID 130269020.
- ↑ i̇Nceoğlu, Özgül; Al-Soud, Waleed Abu; Salles, Joana Falcão; Semenov, Alexander V.; Van Elsas, Jan Dirk (2011). "Comparative Analysis of Bacterial Communities in a Potato Field as Determined by Pyrosequencing". PLOS ONE. 6 (8): e23321. Bibcode:2011PLoSO...623321I. doi:10.1371/journal.pone.0023321. PMC 3158761. PMID 21886785.
- 1 2 3 Bulgarelli, Davide; Rott, Matthias; Schlaeppi, Klaus; Ver Loren Van Themaat, Emiel; Ahmadinejad, Nahal; Assenza, Federica; Rauf, Philipp; Huettel, Bruno; Reinhardt, Richard; Schmelzer, Elmon; Peplies, Joerg; Gloeckner, Frank Oliver; Amann, Rudolf; Eickhorst, Thilo; Schulze-Lefert, Paul (2012). "Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota". Nature. 488 (7409): 91–95. Bibcode:2012Natur.488...91B. doi:10.1038/nature11336. PMID 22859207. S2CID 4393146.
- ↑ Uroz, Stéphane; Buée, Marc; Murat, Claude; Frey-Klett, Pascale; Martin, Francis (2010). "Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil". Environmental Microbiology Reports. 2 (2): 281–288. doi:10.1111/j.1758-2229.2009.00117.x. PMID 23766079.
- ↑ Lundberg, Derek S.; Lebeis, Sarah L.; Paredes, Sur Herrera; Yourstone, Scott; Gehring, Jase; Malfatti, Stephanie; Tremblay, Julien; Engelbrektson, Anna; Kunin, Victor; Rio, Tijana Glavina del; Edgar, Robert C.; Eickhorst, Thilo; Ley, Ruth E.; Hugenholtz, Philip; Tringe, Susannah Green; Dangl, Jeffery L. (2012). "Defining the core Arabidopsis thaliana root microbiome". Nature. 488 (7409): 86–90. Bibcode:2012Natur.488...86L. doi:10.1038/nature11237. PMC 4074413. PMID 22859206.
- 1 2 Schlaeppi, K.; Dombrowski, N.; Oter, R. G.; Ver Loren Van Themaat, E.; Schulze-Lefert, P. (2014). "Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives". Proceedings of the National Academy of Sciences. 111 (2): 585–592. Bibcode:2014PNAS..111..585S. doi:10.1073/pnas.1321597111. PMC 3896156. PMID 24379374. S2CID 13806811.
- ↑ He, Sheng Yang (2020) When plants and their microbes are not in sync, the results can be disastrous The Conversation, 28 August 2020.
- ↑ Lindow, Steven E. (1996). "Role of Immigration and Other Processes in Determining Epiphytic Bacterial Populations". Aerial Plant Surface Microbiology. pp. 155–168. doi:10.1007/978-0-585-34164-4_10. ISBN 978-0-306-45382-3.
- ↑ Friesen, Maren L.; Porter, Stephanie S.; Stark, Scott C.; von Wettberg, Eric J.; Sachs, Joel L.; Martinez-Romero, Esperanza (2011). "Microbially Mediated Plant Functional Traits". Annual Review of Ecology, Evolution, and Systematics. 42: 23–46. doi:10.1146/annurev-ecolsys-102710-145039.
- 1 2 Leveau, Johan HJ (2019). "A brief from the leaf: Latest research to inform our understanding of the phyllosphere microbiome". Current Opinion in Microbiology. 49: 41–49. doi:10.1016/j.mib.2019.10.002. PMID 31707206. S2CID 207946690.
- ↑ Ruinen, J. (1956) "Occurrence of Beijerinckia species in the 'phyllosphere'". Nature, 177(4501): 220–221.
- 1 2 Vogel, Christine; Bodenhausen, Natacha; Gruissem, Wilhelm; Vorholt, Julia A. (2016). "The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health" (PDF). New Phytologist. 212 (1): 192–207. doi:10.1111/nph.14036. hdl:20.500.11850/117578. PMID 27306148.
- 1 2 Cid, Fernanda P.; Maruyama, Fumito; Murase, Kazunori; Graether, Steffen P.; Larama, Giovanni; Bravo, Leon A.; Jorquera, Milko A. (2018). "Draft genome sequences of bacteria isolated from the Deschampsia antarctica phyllosphere". Extremophiles. 22 (3): 537–552. doi:10.1007/s00792-018-1015-x. PMID 29492666. S2CID 4320165.
- ↑ Kumaravel, Sowmya; Thankappan, Sugitha; Raghupathi, Sridar; Uthandi, Sivakumar (2018). "Draft Genome Sequence of Plant Growth-Promoting and Drought-Tolerant Bacillus altitudinis FD48, Isolated from Rice Phylloplane". Genome Announcements. 6 (9). doi:10.1128/genomeA.00019-18. PMC 5834328. PMID 29496824.
- ↑ Laforest‐Lapointe, Isabelle; Whitaker, Briana K. (2019). "Decrypting the phyllosphere microbiota: Progress and challenges". American Journal of Botany. 106 (2): 171–173. doi:10.1002/ajb2.1229. PMID 30726571.
- 1 2 3 4 5 6 7 8 9 10 11 12 Noble, Anya S.; Noe, Stevie; Clearwater, Michael J.; Lee, Charles K. (2020). "A core phyllosphere microbiome exists across distant populations of a tree species indigenous to New Zealand". PLOS ONE. 15 (8): e0237079. Bibcode:2020PLoSO..1537079N. doi:10.1371/journal.pone.0237079. PMC 7425925. PMID 32790769.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.  Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - 1 2 Finkel, Omri M.; Burch, Adrien Y.; Lindow, Steven E.; Post, Anton F.; Belkin, Shimshon (2011). "Geographical Location Determines the Population Structure in Phyllosphere Microbial Communities of a Salt-Excreting Desert Tree". Applied and Environmental Microbiology. 77 (21): 7647–7655. Bibcode:2011ApEnM..77.7647F. doi:10.1128/AEM.05565-11. PMC 3209174. PMID 21926212.
- ↑ Vorholt, Julia A. (2012). "Microbial life in the phyllosphere". Nature Reviews Microbiology. 10 (12): 828–840. doi:10.1038/nrmicro2910. hdl:20.500.11850/59727. PMID 23154261. S2CID 10447146.
- ↑ Lindow, Steven E.; Brandl, Maria T. (2003). "Microbiology of the Phyllosphere". Applied and Environmental Microbiology. 69 (4): 1875–1883. Bibcode:2003ApEnM..69.1875L. doi:10.1128/AEM.69.4.1875-1883.2003. PMC 154815. PMID 12676659. S2CID 2304379.
- ↑ Bodenhausen, Natacha; Horton, Matthew W.; Bergelson, Joy (2013). "Bacterial Communities Associated with the Leaves and the Roots of Arabidopsis thaliana". PLOS ONE. 8 (2): e56329. Bibcode:2013PLoSO...856329B. doi:10.1371/journal.pone.0056329. PMC 3574144. PMID 23457551.
- ↑ Vokou, Despoina; Vareli, Katerina; Zarali, Ekaterini; Karamanoli, Katerina; Constantinidou, Helen-Isis A.; Monokrousos, Nikolaos; Halley, John M.; Sainis, Ioannis (2012). "Exploring Biodiversity in the Bacterial Community of the Mediterranean Phyllosphere and its Relationship with Airborne Bacteria". Microbial Ecology. 64 (3): 714–724. doi:10.1007/s00248-012-0053-7. PMID 22544345. S2CID 17291303.
- ↑ Vorholt, Julia A. (2012). "Microbial life in the phyllosphere". Nature Reviews Microbiology. 10 (12): 828–840. doi:10.1038/nrmicro2910. hdl:20.500.11850/59727. PMID 23154261. S2CID 10447146.
- 1 2 Stone, Bram W. G.; Jackson, Colin R. (2016). "Biogeographic Patterns Between Bacterial Phyllosphere Communities of the Southern Magnolia (Magnolia grandiflora) in a Small Forest". Microbial Ecology. 71 (4): 954–961. doi:10.1007/s00248-016-0738-4. PMID 26883131. S2CID 17292307.
- 1 2 Redford, Amanda J.; Bowers, Robert M.; Knight, Rob; Linhart, Yan; Fierer, Noah (2010). "The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves". Environmental Microbiology. 12 (11): 2885–2893. doi:10.1111/j.1462-2920.2010.02258.x. PMC 3156554. PMID 20545741.
- ↑ Vokou, Despoina; Vareli, Katerina; Zarali, Ekaterini; Karamanoli, Katerina; Constantinidou, Helen-Isis A.; Monokrousos, Nikolaos; Halley, John M.; Sainis, Ioannis (2012). "Exploring Biodiversity in the Bacterial Community of the Mediterranean Phyllosphere and its Relationship with Airborne Bacteria". Microbial Ecology. 64 (3): 714–724. doi:10.1007/s00248-012-0053-7. PMID 22544345. S2CID 17291303.
- 1 2 3 Laforest-Lapointe, Isabelle; Messier, Christian; Kembel, Steven W. (2016). "Host species identity, site and time drive temperate tree phyllosphere bacterial community structure". Microbiome. 4 (1): 27. doi:10.1186/s40168-016-0174-1. PMC 4912770. PMID 27316353.
- ↑ Zarraonaindia, Iratxe; Owens, Sarah M.; Weisenhorn, Pamela; West, Kristin; Hampton-Marcell, Jarrad; Lax, Simon; Bokulich, Nicholas A.; Mills, David A.; Martin, Gilles; Taghavi, Safiyh; Van Der Lelie, Daniel; Gilbert, Jack A. (2015). "The Soil Microbiome Influences Grapevine-Associated Microbiota". mBio. 6 (2). doi:10.1128/mBio.02527-14. PMC 4453523. PMID 25805735.
- ↑ Finkel, Omri M.; Burch, Adrien Y.; Lindow, Steven E.; Post, Anton F.; Belkin, Shimshon (2011). "Geographical Location Determines the Population Structure in Phyllosphere Microbial Communities of a Salt-Excreting Desert Tree". Applied and Environmental Microbiology. 77 (21): 7647–7655. Bibcode:2011ApEnM..77.7647F. doi:10.1128/AEM.05565-11. PMC 3209174. PMID 21926212.
- ↑ Finkel, Omri M.; Burch, Adrien Y.; Elad, Tal; Huse, Susan M.; Lindow, Steven E.; Post, Anton F.; Belkin, Shimshon (2012). "Distance-Decay Relationships Partially Determine Diversity Patterns of Phyllosphere Bacteria on Tamrix Trees across the Sonoran Desert". Applied and Environmental Microbiology. 78 (17): 6187–6193. Bibcode:2012ApEnM..78.6187F. doi:10.1128/AEM.00888-12. PMC 3416633. PMID 22752165.
- 1 2 Kembel, S. W.; O'Connor, T. K.; Arnold, H. K.; Hubbell, S. P.; Wright, S. J.; Green, J. L. (2014). "Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest". Proceedings of the National Academy of Sciences. 111 (38): 13715–13720. Bibcode:2014PNAS..11113715K. doi:10.1073/pnas.1216057111. PMC 4183302. PMID 25225376. S2CID 852584.
- ↑ Innerebner, Gerd; Knief, Claudia; Vorholt, Julia A. (2011). "Protection of Arabidopsis thaliana against Leaf-Pathogenic Pseudomonas syringae by Sphingomonas Strains in a Controlled Model System". Applied and Environmental Microbiology. 77 (10): 3202–3210. Bibcode:2011ApEnM..77.3202I. doi:10.1128/AEM.00133-11. PMC 3126462. PMID 21421777.
- ↑ Lajoie, Geneviève; Maglione, Rémi; Kembel, Steven W. (2020). "Adaptive matching between phyllosphere bacteria and their tree hosts in a neotropical forest". Microbiome. 8 (1): 70. doi:10.1186/s40168-020-00844-7. PMC 7243311. PMID 32438916.
- 1 2 Shade, Ashley; Handelsman, Jo (2012). "Beyond the Venn diagram: The hunt for a core microbiome". Environmental Microbiology. 14 (1): 4–12. doi:10.1111/j.1462-2920.2011.02585.x. PMID 22004523.
- ↑ Berg, Gabriele; Rybakova, Daria; Fischer, Doreen; Cernava, Tomislav; Vergès, Marie-Christine Champomier; Charles, Trevor; Chen, Xiaoyulong; Cocolin, Luca; Eversole, Kellye; Corral, Gema Herrero; Kazou, Maria; Kinkel, Linda; Lange, Lene; Lima, Nelson; Loy, Alexander; MacKlin, James A.; Maguin, Emmanuelle; Mauchline, Tim; McClure, Ryan; Mitter, Birgit; Ryan, Matthew; Sarand, Inga; Smidt, Hauke; Schelkle, Bettina; Roume, Hugo; Kiran, G. Seghal; Selvin, Joseph; Souza, Rafael Soares Correa de; Van Overbeek, Leo; et al. (2020). "Microbiome definition re-visited: Old concepts and new challenges". Microbiome. 8 (1): 103. doi:10.1186/s40168-020-00875-0. PMC 7329523. PMID 32605663.
- ↑ Turnbaugh, Peter J.; Hamady, Micah; Yatsunenko, Tanya; Cantarel, Brandi L.; Duncan, Alexis; Ley, Ruth E.; Sogin, Mitchell L.; Jones, William J.; Roe, Bruce A.; Affourtit, Jason P.; Egholm, Michael; Henrissat, Bernard; Heath, Andrew C.; Knight, Rob; Gordon, Jeffrey I. (2009). "A core gut microbiome in obese and lean twins". Nature. 457 (7228): 480–484. Bibcode:2009Natur.457..480T. doi:10.1038/nature07540. PMC 2677729. PMID 19043404.
- ↑ Lundberg, Derek S.; Lebeis, Sarah L.; Paredes, Sur Herrera; Yourstone, Scott; Gehring, Jase; Malfatti, Stephanie; Tremblay, Julien; Engelbrektson, Anna; Kunin, Victor; Rio, Tijana Glavina del; Edgar, Robert C.; Eickhorst, Thilo; Ley, Ruth E.; Hugenholtz, Philip; Tringe, Susannah Green; Dangl, Jeffery L. (2012). "Defining the core Arabidopsis thaliana root microbiome". Nature. 488 (7409): 86–90. Bibcode:2012Natur.488...86L. doi:10.1038/nature11237. PMC 4074413. PMID 22859206.
- ↑ Hamonts, Kelly; Trivedi, Pankaj; Garg, Anshu; Janitz, Caroline; Grinyer, Jasmine; Holford, Paul; Botha, Frederik C.; Anderson, Ian C.; Singh, Brajesh K. (2018). "Field study reveals core plant microbiota and relative importance of their drivers". Environmental Microbiology. 20 (1): 124–140. doi:10.1111/1462-2920.14031. PMID 29266641. S2CID 10650949.
- ↑ Cernava, Tomislav; Erlacher, Armin; Soh, Jung; Sensen, Christoph W.; Grube, Martin; Berg, Gabriele (2019). "Enterobacteriaceae dominate the core microbiome and contribute to the resistome of arugula (Eruca sativa Mill.)". Microbiome. 7 (1): 13. doi:10.1186/s40168-019-0624-7. PMC 6352427. PMID 30696492.
- ↑ Leff, Jonathan W.; Del Tredici, Peter; Friedman, William E.; Fierer, Noah (2015). "Spatial structuring of bacterial communities within individual Ginkgo bilobatrees". Environmental Microbiology. 17 (7): 2352–2361. doi:10.1111/1462-2920.12695. PMID 25367625.
- ↑ Hernandez-Agreda, Alejandra; Gates, Ruth D.; Ainsworth, Tracy D. (2017). "Defining the Core Microbiome in Corals' Microbial Soup". Trends in Microbiology. 25 (2): 125–140. doi:10.1016/j.tim.2016.11.003. PMID 27919551.
- ↑ Stephens, J. M. C.; Molan, P. C.; Clarkson, B. D. (2005). "A review of Leptospermum scoparium(Myrtaceae) in New Zealand". New Zealand Journal of Botany. 43 (2): 431–449. doi:10.1080/0028825X.2005.9512966. S2CID 53515334.
- ↑ Cooper, R.A.; Molan, P.C.; Harding, K.G. (2002). "The sensitivity to honey of Gram-positive cocci of clinical significance isolated from wounds". Journal of Applied Microbiology. 93 (5): 857–863. doi:10.1046/j.1365-2672.2002.01761.x. PMID 12392533. S2CID 24517001.
- ↑ Rabie, Erika; Serem, June Cheptoo; Oberholzer, Hester Magdalena; Gaspar, Anabella Regina Marques; Bester, Megan Jean (2016). "How methylglyoxal kills bacteria: An ultrastructural study". Ultrastructural Pathology. 40 (2): 107–111. doi:10.3109/01913123.2016.1154914. hdl:2263/52156. PMID 26986806. S2CID 13372064.
- ↑ Adams, Christopher J.; Manley-Harris, Merilyn; Molan, Peter C. (2009). "The origin of methylglyoxal in New Zealand manuka (Leptospermum scoparium) honey". Carbohydrate Research. 344 (8): 1050–1053. doi:10.1016/j.carres.2009.03.020. PMID 19368902.
- ↑ Atrott, Julia; Haberlau, Steffi; Henle, Thomas (2012). "Studies on the formation of methylglyoxal from dihydroxyacetone in Manuka (Leptospermum scoparium) honey". Carbohydrate Research. 361: 7–11. doi:10.1016/j.carres.2012.07.025. PMID 22960208.
- ↑ Mavric, Elvira; Wittmann, Silvia; Barth, Gerold; Henle, Thomas (2008). "Identification and quantification of methylglyoxal as the dominant antibacterial constituent of Manuka (Leptospermum scoparium)honeys from New Zealand". Molecular Nutrition & Food Research. 52 (4): 483–489. doi:10.1002/mnfr.200700282. PMID 18210383.
- ↑ Hamilton, G., Millner, J., Robertson, A. and Stephens, J. (2013) "Assessment of manuka provenances for production of high 'unique manuka factor' honey". Agronomy New Zealand, 43: 139–144.
- ↑ Williams, Simon; King, Jessica; Revell, Maria; Manley-Harris, Merilyn; Balks, Megan; Janusch, Franziska; Kiefer, Michael; Clearwater, Michael; Brooks, Peter; Dawson, Murray (2014). "Regional, Annual, and Individual Variations in the Dihydroxyacetone Content of the Nectar of Ma̅nuka (Leptospermum scoparium) in New Zealand". Journal of Agricultural and Food Chemistry. 62 (42): 10332–10340. doi:10.1021/jf5045958. PMID 25277074.
- ↑ Stephens, J.M.C. (2006) "The factors responsible for the varying levels of UMF® in mānuka (Leptospermum scoparium) honey", Doctoral dissertation, University of Waikato.
- ↑ Noe, Stevie; Manley-Harris, Merilyn; Clearwater, Michael J. (2019). "Floral nectar of wild mānuka (Leptospermum scoparium) varies more among plants than among sites". New Zealand Journal of Crop and Horticultural Science. 47 (4): 282–296. doi:10.1080/01140671.2019.1670681. S2CID 204143940.
- ↑ Nickless, Elizabeth M.; Anderson, Christopher W. N.; Hamilton, Georgie; Stephens, Jonathan M.; Wargent, Jason (2017). "Soil influences on plant growth, floral density and nectar yield in three cultivars of mānuka (Leptospermum scoparium)". New Zealand Journal of Botany. 55 (2): 100–117. doi:10.1080/0028825X.2016.1247732. S2CID 88657399.
- ↑ Clearwater, Michael J.; Revell, Maria; Noe, Stevie; Manley-Harris, Merilyn (2018). "Influence of genotype, floral stage, and water stress on floral nectar yield and composition of mānuka (Leptospermum scoparium)". Annals of Botany. 121 (3): 501–512. doi:10.1093/aob/mcx183. PMC 5838834. PMID 29300875.
- ↑ Johnston, Peter R. (1998). "Leaf endophytes of manuka (Leptospermum scoparium)". Mycological Research. 102 (8): 1009–1016. doi:10.1017/S0953756297005765.
- ↑ McKenzie, E. H. C.; Johnston, P. R.; Buchanan, P. K. (2006). "Checklist of fungi on teatree (Kunzeaand Leptospermumspecies) in New Zealand". New Zealand Journal of Botany. 44 (3): 293–335. doi:10.1080/0028825X.2006.9513025. S2CID 84538904.
- ↑ Wicaksono, Wisnu Adi; Sansom, Catherine E.; Eirian Jones, E.; Perry, Nigel B.; Monk, Jana; Ridgway, Hayley J. (2018). "Arbuscular mycorrhizal fungi associated with Leptospermum scoparium (Mānuka): Effects on plant growth and essential oil content". Symbiosis. 75: 39–50. doi:10.1007/s13199-017-0506-3. S2CID 4819178.
- ↑ Wicaksono, Wisnu Adi; Jones, E. Eirian; Monk, Jana; Ridgway, Hayley J. (2016). "The Bacterial Signature of Leptospermum scoparium (Mānuka) Reveals Core and Accessory Communities with Bioactive Properties". PLOS ONE. 11 (9): e0163717. Bibcode:2016PLoSO..1163717W. doi:10.1371/journal.pone.0163717. PMC 5038978. PMID 27676607.
- 1 2 Vokou, Despoina; Vareli, Katerina; Zarali, Ekaterini; Karamanoli, Katerina; Constantinidou, Helen-Isis A.; Monokrousos, Nikolaos; Halley, John M.; Sainis, Ioannis (2012). "Exploring Biodiversity in the Bacterial Community of the Mediterranean Phyllosphere and its Relationship with Airborne Bacteria". Microbial Ecology. 64 (3): 714–724. doi:10.1007/s00248-012-0053-7. PMID 22544345. S2CID 17291303.
- ↑ Dastogeer, Khondoker M.G.; Li, Hua; Sivasithamparam, Krishnapillai; Jones, Michael G.K.; Du, Xin; Ren, Yonglin; Wylie, Stephen J. (2017). "Metabolic responses of endophytic Nicotiana benthamiana plants experiencing water stress". Environmental and Experimental Botany. 143: 59–71. doi:10.1016/j.envexpbot.2017.08.008.
- ↑ Rodriguez, R. J.; White Jr, J. F.; Arnold, A. E.; Redman, R. S. (2009). "Fungal endophytes: Diversity and functional roles". New Phytologist. 182 (2): 314–330. doi:10.1111/j.1469-8137.2009.02773.x. PMID 19236579.
- ↑ Abdelfattah, Ahmed; Wisniewski, Michael; Schena, Leonardo; Tack, Ayco J.M. (14 May 2020). "Experimental Evidence of Microbial Inheritance in Plants and Transmission Routes from Seed to Phyllosphere and Root". doi:10.21203/rs.3.rs-27656/v1.
{{cite journal}}: Cite journal requires|journal=(help) - ↑ Matsumoto H, Fan X, Wang Y, Kusstatscher P, Duan J, Wu S, et al. (January 2021). "Bacterial seed endophyte shapes disease resistance in rice". Nature Plants. 7 (1): 60–72. doi:10.1038/s41477-020-00826-5. PMID 33398157. S2CID 230508404.
- ↑ "Seeds transfer their microbes to the next generation". EurekAlert!. Retrieved 21 January 2021.
- ↑ Abdelfattah, Ahmed; Wisniewski, Michael; Schena, Leonardo; Tack, Ayco J. M. (2021). "Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root". Environmental Microbiology. 23 (4): 2199–2214. doi:10.1111/1462-2920.15392. ISSN 1462-2920. PMID 33427409.
- ↑ Hassani MA, Durán P, Hacquard S (March 2018). "Microbial interactions within the plant holobiont". Microbiome. 6 (1): 58. doi:10.1186/s40168-018-0445-0. PMC 5870681. PMID 29587885.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Reference books
- Saleem M (2015) Microbiome Community Ecology: Fundamentals and Applications Springer. ISBN 9783319116655.
- Kumar V, Prasad R, Kumar M and Choudhary DK (2019) Microbiome in Plant Health and Disease: Challenges and Opportunities Springer. ISBN 9789811384950.
- Kumar, Vivek; Prasad, Ram; Kumar, Manoj; Choudhary, Devendra K. (10 August 2019). Microbiome in Plant Health and Disease: Challenges and Opportunities. ISBN 9789811384950.
- Grube, Martin; Schloter, Michael; Smalla, Kornelia; Berg, Gabriele (22 January 2015). The plant microbiome and its importance for plant and human health. Frontiers in Microbiology. Vol. 5. p. 491. doi:10.3389/fmicb.2014.00491. ISBN 9782889193783. PMC 4166366. PMID 25278934.
- Egamberdieva, Dilfuza; Ahmad, Parvaiz (6 February 2018). Plant Microbiome: Stress Response. ISBN 9789811055140.
- Castiglione, Stefano; Cicatelli, Angela; Ferrol, Nuria; Rozpadek, Piotr (22 August 2019). Effects of Plant-Microbiome Interactions on Phyto- and Bio-Remediation Capacity. ISBN 9782889459322.
- Castiglione, Stefano; Cicatelli, Angela; Ferrol, Nuria; Rozpadek, Piotr (22 August 2019). Effects of Plant-Microbiome Interactions on Phyto- and Bio-Remediation Capacity. ISBN 9782889459322.
- Carvalhais, Lilia C.; Dennis, Paul G. (30 December 2020). The Plant Microbiome: Methods and Protocols. ISBN 9781071610398.
- Kumar, Vivek; Prasad, Ram; Kumar, Manoj; Choudhary, Devendra K. (10 August 2019). Microbiome in Plant Health and Disease: Challenges and Opportunities. ISBN 9789811384950.
- Solanki, Manoj Kumar; Kashyap, Prem Lal; Ansari, Rizwan Ali; Kumari, Baby (28 August 2020). Microbiomes and Plant Health: Panoply and Their Applications. ISBN 9780128226018.
- Yadav, Ajar (2020). Advances in plant microbiome and sustainable agriculture functional annotation and future challenges. Singapore: Springer. ISBN 978-981-15-3204-7. OCLC 1193118045.
- Varma, Ajit (28 August 2020). Plant Microbiome Paradigm. ISBN 9783030503956.
- Doty, Sharon Lafferty (21 September 2017). Functional Importance of the Plant Microbiome: Implications for Agriculture, Forestry and Bioenergy. ISBN 9783319658971.
- Antwis, Rachael E.; Harrison, Xavier A.; Cox, Michael J. (12 March 2020). Microbiomes of Soils, Plants and Animals: An Integrated Approach. ISBN 9781108473712.
- Varma, Ajit; Tripathi, Swati; Prasad, Ram (20 October 2020). Plant Microbiome Paradigm. ISBN 9783030503949.
- Rosenberg, Eugene; Zilber-Rosenberg, Ilana (31 January 2014). The Hologenome Concept: Human, Animal and Plant Microbiota. ISBN 9783319042411.
- Yadav, Ajar Nath; Singh, Joginder; Rastegari, Ali Asghar; Yadav, Neelam (6 March 2020). Plant Microbiomes for Sustainable Agriculture. ISBN 9783030384531.
| Wikimedia Commons has media related to Plant microbiomes. |