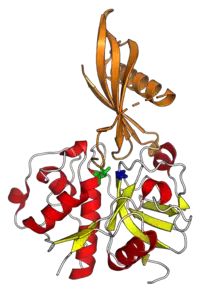
Papain-like protease
Papain-like proteases (or papain-like (cysteine) peptidases; abbreviated PLP or PLCP) are a large protein family of cysteine protease enzymes that share structural and enzymatic properties with the group's namesake member, papain. They are found in all domains of life. In animals, the group is often known as cysteine cathepsins or, in older literature, lysosomal peptidases.[1] In the MEROPS protease enzyme classification system, papain-like proteases form Clan CA.[2] Papain-like proteases share a common catalytic dyad active site featuring a cysteine amino acid residue that acts as a nucleophile.[1]
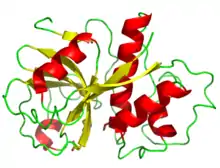 Papain, the family's namesake member, from Carica papaya | |
| Identifiers | |
|---|---|
| Symbol | Peptidase_CA |
| Pfam clan | CL0125 |
| MEROPS | CA |
The human genome encodes eleven cysteine cathepsins which have a broad range of physiological functions.[3] In some parasites papain-like proteases have roles in host invasion, such as cruzipain from Trypanosoma cruzi.[1] In plants, they are involved in host defense and in development.[4] Studies of papain-like proteases from prokaryotes have lagged their eukaryotic counterparts.[1] In cellular organisms they are synthesized as preproenzymes that are not enzymatically active until mature, and their activities are tightly regulated, often by the presence of endogenous protease inhibitors such as cystatins.[3] In many RNA viruses, including significant human pathogens such as the coronaviruses SARS-CoV and SARS-CoV-2, papain-like protease protein domains often have roles in processing of polyproteins into mature viral nonstructural proteins.[5][6] Many papain-like proteases are considered potential drug targets.[3][7]
Classification
The MEROPS system of protease enzyme classification defines clan CA as containing the papain-like proteases. They are thought to have a shared evolutionary origin. As of 2021, the clan contained 45 families.[2][8]
Structure
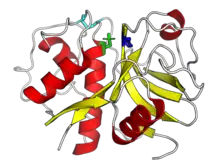
The structure of papain was among the earliest protein structures experimentally determined by X-ray crystallography.[3][10][9] Many papain-like protease enzymes function as monomers, though a few, such as cathepsin C (Dipeptidyl-peptidase I), are homotetramers. The mature monomer structure is characteristically divided into two lobes or subdomains, known as the L-domain (N-terminal) and the R-domain (C-terminal), where the active site is located between them.[1] The L-domain is primarily helical while the R-domain contains beta-sheets in a beta-barrel-like shape, surrounded by a helix.[3] The enzyme substrate interacts with both domains in an extended conformation.[1][3]
Papain-like proteases are often synthesized as preproenzymes, or enzymatically inactive precursors. A signal peptide at the N-terminus, which serves as a subcellular localization signal, is cleaved by signal peptidase to form a zymogen. Post-translational modification in the form of N-linked glycosylation also occurs in parallel.[3] The zymogen is still inactive due to the presence of a propeptide which functions as an inhibitor blocking access to the active site. The propeptide is removed by proteolysis to form the mature enzyme.[1][3][11]
Catalytic mechanism
Papain-like proteases have a catalytic dyad consisting of a cysteine and a histidine residue, which form an ion pair through their charged thiolate and imidazolium side chains. The negatively charged cysteine thiolate functions as a nucleophile.[1][2] Additional neighboring residues - aspartate, asparagine, or glutamine - position the catalytic residues;[1][2] in papain, the required catalytic residues cysteine, histidine, and aspartate are sometimes called the catalytic triad (similar to serine proteases).[11] Papain-like proteases are usually endopeptidases, but some members of the group are also, or even exclusively, exopeptidases.[1] Some viral papain-like proteases, including those of coronaviruses, can also cleave isopeptide bonds and can function as deubiquitinases.[5]
Function
Mammals
In animals, especially in mammalian biology, members of the papain-like protease family are usually referred to as cysteine cathepsins - that is, the cysteine protease members of the group of proteases known as cathepsins (which includes cysteine, serine, and aspartic proteases). In humans, there are 11 cysteine cathepsins: B, C, F, H, K, L, O, S, V, X, and W. Most cathepsins are expressed throughout the body, but some have narrower tissue distribution.[1][3]
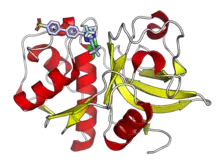
Although historically known as lysosomal proteases and studied mainly for their role in protein catabolism, cysteine cathepsins have since been identified playing major roles in a number of physiological processes and disease states. As part of normal physiological processes, they are involved in key steps of antigen presentation as part of the adaptive immune system, remodeling of the extracellular matrix, differentiation of keratinocytes, and processing of peptide hormones.[1][3] Cysteine cathepsins have been associated with cancer and tumor progression, cardiovascular disease, autoimmune disease, and other human health conditions.[11][13][14] Cathepsin K has a role in bone resorption and has been studied as a drug target for osteoporosis.[15]
Parasites
A number of parasites, including helminths (parasitic worms), use papain-like proteases as mechanisms for invasion of their hosts. Examples include Toxoplasma gondii and Giardia lamblia. In many flatworms, there are very high levels of expression of cysteine cathepsins; in the liver fluke Fasciola hepatica, gene duplications have produced over 20 paralogs of a cathepsin L-like enzyme.[1] Cysteine cathepsins are also part of the normal life cycle of the unicellular parasite Leishmania, where they function as virulence factors.[16] The enzyme and potential drug target cruzipain is important for the life cycle of the parasite Trypanosoma cruzi, which causes Chagas' disease.[17]
Plants
Members of the papain-like protease family play a number of important roles in plant development, including seed germination, leaf senescence, and responding to abiotic stress. Papain-like proteases are involved in regulation of programmed cell death in plants, for example in tapetum during development of pollen. They are also important in plant immunity providing defense against pests and pathogens.[4] The relationship between plant papain-like proteases and pathogen responses - such as cystatin inhibitors - have been described as an evolutionary arms race.[19]
Some PLP family members in plants have culinary and commercial applications. The family's namesake member, papain, is a protease derived from papaya, used as a meat tenderizer.[20] Similar but less widely used plant products include bromelain from pineapple and ficin from figs.[1][20]
Prokaryotes
Although papain-like proteases are found in all domains of life, they have been less well-studied in prokaryotes than in eukaryotes.[1] Only a few prokaryotic PLP enzymes have been characterized by X-ray crystallography or enzymatic studies, mostly from pathogenic bacteria, including streptopain from Streptococcus pyogenes; xylellain, from the plant pathogen Xylella fastidiosa;[21] Cwp84 from Clostridium difficile;[22] and Lpg2622 from Legionella pneumophila.[23]
Viruses
The papain-like protease family includes a number of protein domains that are found in large polyproteins expressed by RNA viruses.[2] Among the best studied viral PLPs are nidoviral papain-like protease domains from nidoviruses, particularly those from coronaviruses. These PLPs are responsible for several cleavage events that process a large polyprotein into viral nonstructural proteins, although they perform fewer cleavages than the 3C-like protease (also known as the main protease).[5] Coronavirus PLPs are multifunctional enzymes that can also act as deubiquitinases (cleaving the isopeptide bond to ubiquitin) and "deISGylating enzymes" with analogous activity against the ubiquitin-like protein ISG15.[5][6] In human pathogens including SARS-CoV, MERS-CoV, and SARS-CoV-2, the PLP domain is essential for viral replication and is therefore considered a drug target for the development of antiviral drugs.[6][7]
References
- Novinec, Marko; Lenarčič, Brigita (1 June 2013). "Papain-like peptidases: structure, function, and evolution". BioMolecular Concepts. 4 (3): 287–308. doi:10.1515/bmc-2012-0054. PMID 25436581. S2CID 2112616.
- "Summary for clan CA". MEROPS. Retrieved 20 December 2021.
- Turk, Vito; Stoka, Veronika; Vasiljeva, Olga; Renko, Miha; Sun, Tao; Turk, Boris; Turk, Dušan (January 2012). "Cysteine cathepsins: From structure, function and regulation to new frontiers". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1824 (1): 68–88. doi:10.1016/j.bbapap.2011.10.002. PMC 7105208. PMID 22024571.
- Liu, Huijuan; Hu, Menghui; Wang, Qi; Cheng, Lin; Zhang, Zaibao (4 December 2018). "Role of Papain-Like Cysteine Proteases in Plant Development". Frontiers in Plant Science. 9: 1717. doi:10.3389/fpls.2018.01717. PMC 6288466. PMID 30564252.
- Mielech, Anna M.; Chen, Yafang; Mesecar, Andrew D.; Baker, Susan C. (December 2014). "Nidovirus papain-like proteases: Multifunctional enzymes with protease, deubiquitinating and deISGylating activities". Virus Research. 194: 184–190. doi:10.1016/j.virusres.2014.01.025. PMC 4125544. PMID 24512893.
- Hartenian, Ella; Nandakumar, Divya; Lari, Azra; Ly, Michael; Tucker, Jessica M.; Glaunsinger, Britt A. (September 2020). "The molecular virology of coronaviruses". Journal of Biological Chemistry. 295 (37): 12910–12934. doi:10.1074/jbc.REV120.013930. PMC 7489918. PMID 32661197.
- Klemm, Theresa; Ebert, Gregor; Calleja, Dale J; Allison, Cody C; Richardson, Lachlan W; Bernardini, Jonathan P; Lu, Bernadine GC; Kuchel, Nathan W; Grohmann, Christoph; Shibata, Yuri; Gan, Zhong Yan; Cooney, James P; Doerflinger, Marcel; Au, Amanda E; Blackmore, Timothy R; Heden van Noort, Gerbrand J; Geurink, Paul P; Ovaa, Huib; Newman, Janet; Riboldi‐Tunnicliffe, Alan; Czabotar, Peter E; Mitchell, Jeffrey P; Feltham, Rebecca; Lechtenberg, Bernhard C; Lowes, Kym N; Dewson, Grant; Pellegrini, Marc; Lessene, Guillaume; Komander, David (15 September 2020). "Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2". The EMBO Journal. 39 (18): e106275. doi:10.15252/embj.2020106275. PMC 7461020. PMID 32845033. S2CID 221328909.
- Rawlings, Neil D; Barrett, Alan J; Thomas, Paul D; Huang, Xiaosong; Bateman, Alex; Finn, Robert D (4 January 2018). "The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database". Nucleic Acids Research. 46 (D1): D624–D632. doi:10.1093/nar/gkx1134. PMC 5753285. PMID 29145643.
- Kamphuis, I.G.; Kalk, K.H.; Swarte, M.B.A.; Drenth, J. (October 1984). "Structure of papain refined at 1.65 Å resolution". Journal of Molecular Biology. 179 (2): 233–256. doi:10.1016/0022-2836(84)90467-4. PMID 6502713.
- Drenth, J.; Jansonius, J. N.; Koekoek, R.; Swen, H. M.; Wolthers, B. G. (June 1968). "Structure of Papain". Nature. 218 (5145): 929–932. Bibcode:1968Natur.218..929D. doi:10.1038/218929a0. PMID 5681232. S2CID 4169127.
- Löser, Reik; Pietzsch, Jens (23 June 2015). "Cysteine cathepsins: their role in tumor progression and recent trends in the development of imaging probes". Frontiers in Chemistry. 3: 37. Bibcode:2015FrCh....3...37L. doi:10.3389/fchem.2015.00037. PMC 4477214. PMID 26157794.
- Law, Simon; Andrault, Pierre-Marie; Aguda, Adeleke H.; Nguyen, Nham T.; Kruglyak, Natasha; Brayer, Gary D.; Brömme, Dieter (1 March 2017). "Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib". Biochemical Journal. 474 (5): 851–864. doi:10.1042/BCJ20160985. PMID 28049758.
- Vidak, Eva; Javoršek, Urban; Vizovišek, Matej; Turk, Boris (20 March 2019). "Cysteine Cathepsins and their Extracellular Roles: Shaping the Microenvironment". Cells. 8 (3): 264. doi:10.3390/cells8030264. PMC 6468544. PMID 30897858.
- Jakoš, Tanja; Pišlar, Anja; Jewett, Anahid; Kos, Janko (28 August 2019). "Cysteine Cathepsins in Tumor-Associated Immune Cells". Frontiers in Immunology. 10: 2037. doi:10.3389/fimmu.2019.02037. PMC 6724555. PMID 31555270.
- Drake, Matthew T; Clarke, Bart L; Oursler, Merry Jo; Khosla, Sundeep (1 August 2017). "Cathepsin K Inhibitors for Osteoporosis: Biology, Potential Clinical Utility, and Lessons Learned". Endocrine Reviews. 38 (4): 325–350. doi:10.1210/er.2015-1114. PMC 5546879. PMID 28651365.
- Mottram, Jeremy C; Coombs, Graham H; Alexander, James (August 2004). "Cysteine peptidases as virulence factors of Leishmania". Current Opinion in Microbiology. 7 (4): 375–381. doi:10.1016/j.mib.2004.06.010. PMID 15358255.
- Branquinha, M. H.; Oliveira, S. S. C.; Sangenito, L. S.; Sodre, C. L.; Kneipp, L. F.; d’Avila-Levy, C. M.; Santos, A. L. S. (2015). "Cruzipain: An Update on its Potential as Chemotherapy Target against the Human Pathogen Trypanosoma cruzi". Current Medicinal Chemistry. 22 (18): 2225–2235. doi:10.2174/0929867322666150521091652. PMID 25994861.
- Chu, Ming-Hung; Liu, Kai-Lun; Wu, Hsin-Yi; Yeh, Kai-Wun; Cheng, Yi-Sheng (August 2011). "Crystal structure of tarocystatin–papain complex: implications for the inhibition property of group-2 phytocystatins". Planta. 234 (2): 243–254. doi:10.1007/s00425-011-1398-8. PMC 3144364. PMID 21416241.
- Misas‐Villamil, Johana C.; Hoorn, Renier A. L.; Doehlemann, Gunther (December 2016). "Papain‐like cysteine proteases as hubs in plant immunity". New Phytologist. 212 (4): 902–907. doi:10.1111/nph.14117. PMID 27488095.
- Fernández-Lucas, Jesús; Castañeda, Daniel; Hormigo, Daniel (October 2017). "New trends for a classical enzyme: Papain, a biotechnological success story in the food industry". Trends in Food Science & Technology. 68: 91–101. doi:10.1016/j.tifs.2017.08.017. hdl:11323/1609.
- Leite, Ney Ribeiro; Faro, Aline Regis; Dotta, Maria Amélia Oliva; Faim, Livia Maria; Gianotti, Andreia; Silva, Flavio Henrique; Oliva, Glaucius; Thiemann, Otavio Henrique (14 February 2013). "The crystal structure of the cysteine protease Xylellain from Xylella fastidiosa reveals an intriguing activation mechanism". FEBS Letters. 587 (4): 339–344. doi:10.1016/j.febslet.2013.01.009. PMID 23333295. S2CID 1367730.
- Bradshaw, William J.; Kirby, Jonathan M.; Thiyagarajan, Nethaji; Chambers, Christopher J.; Davies, Abigail H.; Roberts, April K.; Shone, Clifford C.; Acharya, K. Ravi (1 July 2014). "The structure of the cysteine protease and lectin-like domains of Cwp84, a surface layer-associated protein from Clostridium difficile". Acta Crystallographica Section D: Biological Crystallography. 70 (7): 1983–1993. doi:10.1107/S1399004714009997. PMC 4089489. PMID 25004975.
- Gong, Xiaojian; Zhao, Xiaolei; Zhang, Wei; Wang, Jinzhao; Chen, Xiaofang; Hameed, Muhammad Fazal; Zhang, Nannan; Ge, Honghua (August 2018). "Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila". FEBS Letters. 592 (16): 2798–2810. doi:10.1002/1873-3468.13210. PMID 30071124. S2CID 51906615.
- Osipiuk, Jerzy; Azizi, Saara-Anne; Dvorkin, Steve; Endres, Michael; Jedrzejczak, Robert; Jones, Krysten A.; Kang, Soowon; Kathayat, Rahul S.; Kim, Youngchang; Lisnyak, Vladislav G.; Maki, Samantha L.; Nicolaescu, Vlad; Taylor, Cooper A.; Tesar, Christine; Zhang, Yu-An; Zhou, Zhiyao; Randall, Glenn; Michalska, Karolina; Snyder, Scott A.; Dickinson, Bryan C.; Joachimiak, Andrzej (December 2021). "Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors". Nature Communications. 12 (1): 743. Bibcode:2021NatCo..12..743O. doi:10.1038/s41467-021-21060-3. PMC 7854729. PMID 33531496.