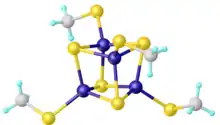
Iron–sulfur cluster
Iron–sulfur clusters are molecular ensembles of iron and sulfide. They are most often discussed in the context of the biological role for iron–sulfur proteins, which are pervasive.[2] Many Fe–S clusters are known in the area of organometallic chemistry and as precursors to synthetic analogues of the biological clusters (see Figure). It is believed that the last universal common ancestor had many iron-sulfur clusters.[3]
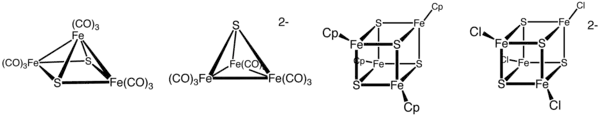
Organometallic clusters
Organometallic Fe–S clusters include the sulfido carbonyls with the formula Fe2S2(CO)6, H2Fe3S(CO)9, and Fe3S2(CO)9. Compounds are also known that incorporate cyclopentadienyl ligands, such as (C5H5)4Fe4S4.[4]
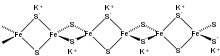
Inorganic materials
Biological Fe–S clusters
Iron–sulfur clusters occur in many biological systems, often as components of electron transfer proteins. The ferredoxin proteins are the most common Fe–S clusters in nature. They feature either 2Fe–2S or 4Fe–4S centers. They occur in all branches of life.[5]
Fe–S clusters can be classified according to their Fe:S stoichiometry [2Fe–2S], [4Fe–3S], [3Fe–4S], and [4Fe–4S].[6] The [4Fe–4S] clusters occur in two forms: normal ferredoxins and high potential iron proteins (HiPIP). Both adopt cuboidal structures, but they utilize different oxidation states. They are found in all forms of life.[7]
The relevant redox couple in all Fe–S proteins is Fe(II)/Fe(III).[7]
Many clusters have been synthesized in the laboratory with the formula [Fe4S4(SR)4]2−, which are known for many R substituents, and with many cations. Variations have been prepared including the incomplete cubanes [Fe3S4(SR)3]3−.[8]
The Rieske proteins contain Fe–S clusters that coordinate as a 2Fe–2S structure and can be found in the membrane bound cytochrome bc1 complex III in the mitochondria of eukaryotes and bacteria. They are also a part of the proteins of the chloroplast such as the cytochrome b6f complex in photosynthetic organisms. These photosynthetic organisms include plants, green algae, and cyanobacteria, the bacterial precursor to chloroplasts. Both are part of the electron transport chain of their respective organisms which is a crucial step in the energy harvesting for many organisms.[9]
In some instances Fe–S clusters are redox-inactive, but are proposed to have structural roles. Examples include endonuclease III and MutY.[5][10]
See also
References
- Axel Kern; Christian Näther; Felix Studt; Felix Tuczek (2004). "Application of a Universal Force Field to Mixed Fe/Mo−S/Se Cubane and Heterocubane Clusters. 1. Substitution of Sulfur by Selenium in the Series [Fe4X4(YCH3)4]2-; X = S/Se and Y = S/Se". Inorg. Chem. 43 (16): 5003–5010. doi:10.1021/ic030347d. PMID 15285677.
- S. J. Lippard, J. M. Berg "Principles of Bioinorganic Chemistry" University Science Books: Mill Valley, CA; 1994. ISBN 0-935702-73-3.
- Weiss, Madeline C., et al. "The physiology and habitat of the last universal common ancestor." Nature microbiology 1.9 (2016): 1-8.
- Ogino, H.; Inomata, S.; Tobita, H. (1998). "Abiological Iron-Sulfur Clusters". Chem. Rev. 98 (6): 2093–2122. doi:10.1021/cr940081f. PMID 11848961.
- Johnson, D. C.; Dean, D. R.; Smith, A. D.; Johnson, M. K. (2005). "Structure, function, and formation of biological iron-sulfur clusters". Annual Review of Biochemistry. 74: 247–281. doi:10.1146/annurev.biochem.74.082803.133518. PMID 15952888.
- Lill, Roland (2015). "Issue of iron-sulfur protein". Biochimica et Biophysica Acta. 1853 (6): 1251–1252. doi:10.1016/j.bbamcr.2015.03.001. PMC 5501863. PMID 25746719.
- Fisher, N (1998). "Intramolecular electron transfer in [4Fe–4S)]". The EMBO Journal: 849–858.
- Rao, P. V.; Holm, R. H. (2004). "Synthetic Analogues of the Active Sites of Iron-Sulfur Proteins". Chem. Rev. 104 (2): 527─559. doi:10.1021/Cr020615+. PMID 14871134.
- BIOLOGICAL INORGANIC CHEMISTRY: structure and reactivity. [S.l.]: UNIVERSITY SCIENCE BOOKS. 2018. ISBN 978-1-938787-96-6. OCLC 1048090793.
- Guan, Y.; Manuel, R. C.; Arvai, A. S.; Parikh, S. S.; Mol, C. D.; Miller, J. H.; Lloyd, S.; Tainer, J. A. (December 1998). "MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily". Nature Structural Biology. 5 (12): 1058–1064. doi:10.1038/4168. ISSN 1072-8368. PMID 9846876. S2CID 22085836.