Asgard (archaea)
Asgard or Asgardarchaeota[2] is a proposed superphylum consisting of a group of archaea that includes Lokiarchaeota, Thorarchaeota, Odinarchaeota, and Heimdallarchaeota.[3] It appears the eukaryotes have emerged within the Asgard, in a branch containing the Heimdallarchaeota.[4][5] This supports the two-domain system of classification over the three-domain system.[6][7]
| Asgard | |
|---|---|
| Scientific classification | |
| Domain: | Archaea |
| Kingdom: | Proteoarchaeota |
| Superphylum: | Asgard Katarzyna Zaremba-Niedzwiedzka, et al. 2017 |
| Phyla | |
 | |
| Synonyms | |
| |
Discovery and Nomenclature
In the summer of 2010, sediments from a gravity core taken in the rift valley on the Knipovich ridge in the Arctic Ocean, near the so-called Loki's Castle hydrothermal vent site, were analysed. Specific sediment horizons previously shown to contain high abundances of novel archaeal lineages, were subjected to metagenomic analysis.[8][9]
In 2015, an Uppsala University-led team proposed the Lokiarchaeota phylum based on phylogenetic analyses using a set of highly conserved protein-coding genes.[10] Through a reference to the hydrothermal vent complex from which the first genome sample originated, the name refers to Loki, the Norse shape-shifting god.[11] The Loki of mythology has been described as "a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies",[12] analogous to the role of Lokiarchaeota in the debates about the origin of eukaryotes.[10][13]
In 2016, a University of Texas-led team discovered Thorarchaeota from samples taken from the White Oak River in North Carolina, named in reference to Thor, another Norse god.[14]
Additional samples from Loki's Castle, Yellowstone National Park, Aarhus Bay, an aquifer near the Colorado River, New Zealand's Radiata Pool, hydrothermal vents near Taketomi Island, Japan, and the White Oak River estuary in the United States led researchers to discover Odinarchaeota and Heimdallarchaeota,[3] and following the naming convention having been established to use Norse deities, the archaea were named for Odin and Heimdallr, respectively. Researchers therefore, named the superphylum containing these microbes “Asgard”, after the realm of the deities in Norse mythology.[3]
Description
Asgard members encode many eukaryotic signature proteins, including novel GTPases, membrane-remodelling proteins like ESCRT and SNF7, a ubiquitin modifier system, and N-glycosylation pathway homologs.[3]
Asgard archaeons have a regulated actin cytoskeleton, and the profilins and gelsolins they use can interact with eukaryotic actins.[15][16] In addition, Asgard archaea tubulin from hydrothermal-living Odinarchaeota (OdinTubulin) was identified as a genuine tubulin. OdinTubulin forms protomers and protofilaments most similar to eukaryotic microtubules, yet assembles into ring systems more similar to FtsZ, indicating that OdinTubulin may represent an evolution intermediate between FtsZ and microtubule-forming tubulins.[17] They also seem to form vesicles under cryogenic electron microscopy. Some may have a PKD domain S-layer.[18] They also share the three-way ES39 expansion in LSU rRNA with eukaryotes.[19]
Metabolism
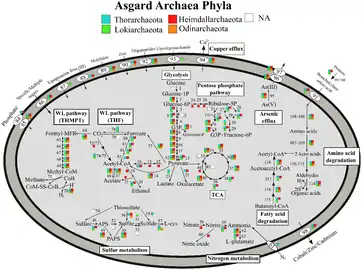 Metabolic pathways of Asgard archaea, variation by Phyla[20]
Metabolic pathways of Asgard archaea, variation by Phyla[20]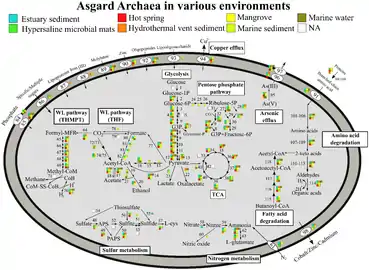 Metabolic pathways of Asgard archaea, variation by environment[20]
Metabolic pathways of Asgard archaea, variation by environment[20]
Asgard archaea are generally obligate anaerobes, though Kariarchaeota, Gerdarchaeota and Hodarchaeota may be facultative aerobes.[21] They have a Wood–Ljungdahl pathway and perform glycolysis. Members can be autotrophs, heterotrophs, or phototrophs using heliorhodopsin.[20] One member, Candidatus Prometheoarchaeum syntrophicum, performs syntrophy with a sulfur-reducing proteobacteria and a methanogenic archaea.[18]
The RuBisCO they have are not carbon-fixing, but likely used for nucleoside salvaging.[20]
Eukaryotic-like features in subdivisions
The phylum "Heimdallarchaeota" was found to have N-terminal core histone tails, a feature previously thought to be exclusively eukaryotic, in 2017. Two other archaeal phyla, both outside of Asgard, were found to also have tails in 2018.[22]
In January 2020, scientists found Candidatus Prometheoarchaeum syntrophicum, a member of Lokiarcheota, engaging in cross-feeding with two bacterial species. Drawing an analogy to symbiogenesis, they consider this relationship a possible link between the simple prokaryotic microorganisms and the complex eukaryotic microorganisms occurring approximately two billion years ago.[23][18]
Classification
The phylogenetic relationship of this group is still under discussion.
| Williams et al. 2019,[5] Eme et al. 2017,[4] Liu et al. 2021[24] & Liu et al. 2020[21] | 53 marker proteins based GTDB 07-RS207[25][26][27] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Taxonomy
In the depicted scenario, the Eukaryota are deep in the tree of Asgard, and any of the Eukaryota is significantly closer related to the Heimdalarchaeota than e.g. the Lokiarchaeota are.
Some authors have suggested to split Heimdallarchaeota into multiple groups (Hodarchaeota, Gerdarchaeota, Kariarchaeota and Heimdallarchaeota). Eukaryotes may be sister to the previous four groups and Wukongarchaeota, or to the entire Asgard archaea group. A favored scenario is syntrophy, where one organism depends on the feeding of the other. In this case, the syntrophy may have been due to the Asgard archaea having been incorporated in an unknown type of bacteria, developing into the nucleus. An α-proteobacterium was incorporated to become the mitochondrion.[28]
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[29] and National Center for Biotechnology Information (NCBI).[30]
- Phylum ?"Freyrarchaeota" Caceres 2019 ex Xie et al. 2021
- Phylum ?"Friggarchaeota" Caceres 2019
- Phylum ?"Gefionarchaeota" Caceres 2019
- Phylum ?"Idunnarchaeota" Caceres 2019
- Phylum ?"Njordarchaeota" Xie et al. 2021
- Phylum ?"Sigynarchaeota" Xie et al. 2021
- Phylum ?"Tyrarchaeota" Xie et al. 2021
- Class "Sifarchaeia" Sun et al. 2021
- Order "Borrarchaeales" Liu et al. 2020
- Family "Borrarchaeaceae" Liu et al. 2020
- "Candidatus Borrarchaeum" Liu et al. 2020
- Family "Borrarchaeaceae" Liu et al. 2020
- Order "Sifarchaeales" Sun et al. 2021
- Family "Sifarchaeaceae" Sun et al. 2021
- "Candidatus Sifarchaeum" corrig. Farag, Zhao & Biddle 2020
- Family "Sifarchaeaceae" Sun et al. 2021
- Order "Borrarchaeales" Liu et al. 2020
- Class "Wukongarchaeia" Liu et al. 2020
- Order "Wukongarchaeales" Liu et al. 2020
- Family "Wukongarchaeaceae" Liu et al. 2020
- "Candidatus Wukongarchaeum" Liu et al. 2020
- Family "Wukongarchaeaceae" Liu et al. 2020
- Order "Wukongarchaeales" Liu et al. 2020
- Class "Heimdallarchaeia"
- Order "Gerdarchaeales" (JABLTI01)
- Order "Heimdallarchaeales"
- Order "Hodarchaeales" Liu et al. 2020
- Family "Hodarchaeaceae" Liu et al. 2020
- "Candidatus Hodarchaeum" Liu et al. 2020
- Family "Hodarchaeaceae" Liu et al. 2020
- Order "Kariarchaeales" Liu et al. 2020
- Family "Kariarchaeaceae" Liu et al. 2020
- "Candidatus Kariarchaeum" Liu et al. 2020
- Family "Kariarchaeaceae" Liu et al. 2020
- Class "Jordarchaeia" Sun et al. 2021
- Order "Jordarchaeales" Sun et al. 2021
- Family "Jordarchaeaceae" Sun et al. 2021
- "Candidatus Jordarchaeum" Sun et al. 2021
- Family "Jordarchaeaceae" Sun et al. 2021
- Order "Jordarchaeales" Sun et al. 2021
- Class "Odinarchaeia" Tamarit et al. 2022
- Order "Odinarchaeales" Tamarit et al. 2022
- Family "Odinarchaeaceae" Tamarit et al. 2022
- "Candidatus Odinarchaeum" Tamarit et al. 2022
- Family "Odinarchaeaceae" Tamarit et al. 2022
- Order "Odinarchaeales" Tamarit et al. 2022
- Class "Baldrarchaeia" Liu et al. 2020
- Order "Baldrarchaeales" Liu et al. 2020
- Family "Baldrarchaeaceae" Liu et al. 2020
- "Candidatus Baldrarchaeum" Liu et al. 2020
- Family "Baldrarchaeaceae" Liu et al. 2020
- Order "Baldrarchaeales" Liu et al. 2020
- Class "Thorarchaeia"
- Order "Thorarchaeales"
- Family "Thorarchaeaceae" (MBG-B)
- Order "Thorarchaeales"
- Class "Hermodarchaeia" Liu et al. 2020
- Order "Hermodarchaeales" Liu et al. 2020
- Family "Hermodarchaeaceae" Liu et al. 2020
- "Candidatus Hermodarchaeum" Liu et al. 2020
- Family "Hermodarchaeaceae" Liu et al. 2020
- Order "Hermodarchaeales" Liu et al. 2020
- Class "Lokiarchaeia" corrig. Spang et al. 2015
- Order "Helarchaeales"
- Order "Lokiarchaeales" Spang et al. 2015
- Family "Lokiarchaeaceae" Vanwonterghem et al. 2016
- "Candidatus Lokiarchaeum" corrig. Spang et al. 2015 (MBGB, DSAG)
- Family "MK-D1"
- "Candidatus Promethearchaeum" corrig. Imachi, Nobu & Takai 2020
- Family "Lokiarchaeaceae" Vanwonterghem et al. 2016
Asgard archaeal mobilome
Viruses
Several family-level groups of viruses associated with Asgard archaea have been discovered using metagenomics.[31][32][33] The viruses were assigned to Lokiarchaeia, Thorarchaeia, Odinarchaeia and Helarchaeia hosts using CRISPR spacer matching to the corresponding protospacers within the viral genomes. Two groups of viruses are related to arcaheal and bacterial viruses of the class Caudoviricetes, i.e., viruses with icosahedral capsids and helical tails;[31][33] two other distinct groups are distantly related to tailless archaeal and bacterial viruses with icosahedral capsids of the realm Varidnaviria;[31][32] and the third group of viruses is related to archaea-specific viruses with lemon-shaped virus particles.[31][32] The viruses have been identified in deep-sea sediments[31][33] and a terrestrial hot spring of the Yellowstone National Park.[32] All these viruses display very low sequence similarity to other known viruses but are generally related to the previously described prokaryotic viruses,[34] with no meaningful affinity to viruses of eukaryotes.[35][31]
Mobile genetic elements
In addition to viruses, several groups of cryptic mobile genetic elements (MGE) have been discovered through CRISPR spacer matching to be associated with Asgard archaea of the Lokiarchaeia, Thorarchaeia and Heimdallarchaeia lineages.[31][36] These MGE do not encode recognizable viral hallmark proteins and could represent either novel types of viruses or plasmids.
See also
References
- Fournier GP, Poole AM (2018). "A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree". Frontiers in Microbiology. 9: 1896. doi:10.3389/fmicb.2018.01896. PMC 6104171. PMID 30158917.
- Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P (June 2017). "Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes". PLOS Genetics. 13 (6): e1006810. doi:10.1371/journal.pgen.1006810. PMC 5484517. PMID 28604769.
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, et al. (January 2017). "Asgard archaea illuminate the origin of eukaryotic cellular complexity". Nature. 541 (7637): 353–358. Bibcode:2017Natur.541..353Z. doi:10.1038/nature21031. OSTI 1580084. PMID 28077874. S2CID 4458094.
- Eme L, Spang A, Lombard J, Stairs CW, Ettema TJ (November 2017). "Archaea and the origin of eukaryotes". Nature Reviews. Microbiology. 15 (12): 711–723. doi:10.1038/nrmicro.2017.133. PMID 29123225. S2CID 8666687.
- Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM (January 2020). "Phylogenomics provides robust support for a two-domains tree of life". Nature Ecology & Evolution. 4 (1): 138–147. doi:10.1038/s41559-019-1040-x. PMC 6942926. PMID 31819234.
- Nobs SJ, MacLeod FI, Wong HL, Burns BP (May 2022). "Eukarya the chimera: eukaryotes, a secondary innovation of the two domains of life?". Trends in Microbiology. 30 (5): 421–431. doi:10.1016/j.tim.2021.11.003. PMID 34863611. S2CID 244823103.
- Doolittle WF (February 2020). "Evolution: Two Domains of Life or Three?". Current Biology. 30 (4): R177–R179. doi:10.1016/j.cub.2020.01.010. PMID 32097647.
- Jorgensen SL, Hannisdal B, Lanzén A, Baumberger T, Flesland K, Fonseca R, et al. (October 2012). "Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge". Proceedings of the National Academy of Sciences of the United States of America. 109 (42): E2846–E2855. doi:10.1073/pnas.1207574109. PMC 3479504. PMID 23027979.
- Jørgensen SL, Thorseth IH, Pedersen RB, Baumberger T, Schleper C (October 4, 2013). "Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge". Frontiers in Microbiology. 4: 299. doi:10.3389/fmicb.2013.00299. PMC 3790079. PMID 24109477.
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, et al. (May 2015). "Complex archaea that bridge the gap between prokaryotes and eukaryotes". Nature. 521 (7551): 173–179. Bibcode:2015Natur.521..173S. doi:10.1038/nature14447. PMC 4444528. PMID 25945739.
- Yong E. "Break in the Search for the Origin of Complex Life". The Atlantic. Retrieved 2018-03-21.
- von Schnurbein S (November 2000). "The Function of Loki in Snorri Sturluson's "Edda"". History of Religions. 40 (2): 109–124. doi:10.1086/463618.
- Spang A, Eme L, Saw JH, Caceres EF, Zaremba-Niedzwiedzka K, Lombard J, et al. (March 2018). "Asgard archaea are the closest prokaryotic relatives of eukaryotes". PLOS Genetics. 14 (3): e1007080. doi:10.1371/journal.pgen.1007080. PMC 5875740. PMID 29596421.
- Seitz KW, Lazar CS, Hinrichs KU, Teske AP, Baker BJ (July 2016). "Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction". The ISME Journal. 10 (7): 1696–1705. doi:10.1038/ismej.2015.233. PMC 4918440. PMID 26824177.
- Akıl C, Robinson RC (October 2018). "Genomes of Asgard archaea encode profilins that regulate actin". Nature. 562 (7727): 439–443. Bibcode:2018Natur.562..439A. doi:10.1038/s41586-018-0548-6. PMID 30283132. S2CID 52917038.
- Akıl C, Tran LT, Orhant-Prioux M, Baskaran Y, Manser E, Blanchoin L, Robinson RC (August 2020). "Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea". Proceedings of the National Academy of Sciences of the United States of America. 117 (33): 19904–19913. bioRxiv 10.1101/768580. doi:10.1073/pnas.2009167117. PMC 7444086. PMID 32747565.
- Akıl C, Ali S, Tran LT, Gaillard J, Li W, Hayashida K, et al. (March 2022). "Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution". Science Advances. 8 (12): eabm2225. Bibcode:2022SciA....8M2225A. doi:10.1126/sciadv.abm2225. PMC 8956254. PMID 35333570.
- Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, et al. (January 2020). "Isolation of an archaeon at the prokaryote-eukaryote interface". Nature. 577 (7791): 519–525. Bibcode:2020Natur.577..519I. doi:10.1038/s41586-019-1916-6. PMC 7015854. PMID 31942073.
- Penev PI, Fakhretaha-Aval S, Patel VJ, Cannone JJ, Gutell RR, Petrov AS, et al. (October 2020). "Supersized Ribosomal RNA Expansion Segments in Asgard Archaea". Genome Biology and Evolution. 12 (10): 1694–1710. doi:10.1093/gbe/evaa170. PMC 7594248. PMID 32785681.
- MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP (2019). "Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes". AIMS Microbiology. 5 (1): 48–61. doi:10.3934/microbiol.2019.1.48. PMC 6646929. PMID 31384702.
- Liu Y, Makarova KS, Huang WC, Wolf YI, Nikolskaya A, Zhang X, et al. (2020). "Expanding diversity of Asgard archaea and the elusive ancestry of eukaryotes". bioRxiv. doi:10.1101/2020.10.19.343400. S2CID 225056970.
- Henneman B, van Emmerik C, van Ingen H, Dame RT (September 2018). "Structure and function of archaeal histones". PLOS Genetics. 14 (9): e1007582. Bibcode:2018BpJ...114..446H. doi:10.1371/journal.pgen.1007582. PMC 6136690. PMID 30212449.
- Zimmer C (15 January 2020). "This Strange Microbe May Mark One of Life's Great Leaps - A organism living in ocean muck offers clues to the origins of the complex cells of all animals and plants". The New York Times. Retrieved 16 January 2020.
- Liu Y, Makarova KS, Huang WC, Wolf YI, Nikolskaya AN, Zhang X, et al. (May 2021). "Expanded diversity of Asgard archaea and their relationships with eukaryotes". Nature. 593 (7860): 553–557. Bibcode:2021Natur.593..553L. doi:10.1038/s41586-021-03494-3. PMID 33911286. S2CID 233447651.
- "GTDB release 07-RS207". Genome Taxonomy Database. Retrieved 6 December 2021.
- "ar53_r207.sp_label". Genome Taxonomy Database. Retrieved 20 June 2021.
- "Taxon History". Genome Taxonomy Database. Retrieved 6 December 2021.
- López-García P, Moreira D (July 2019). "Eukaryogenesis, a syntrophy affair". Nature Microbiology. 4 (7): 1068–1070. doi:10.1038/s41564-019-0495-5. PMC 6684364. PMID 31222170.
- Euzéby JP. "Superphylum "Asgardarchaeota"". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2021-06-27.
- Sayers; et al. "Asgard group". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2021-03-20.
- Medvedeva S, Sun J, Yutin N, Koonin EV, Nunoura T, Rinke C, Krupovic M (July 2022). "Three families of Asgard archaeal viruses identified in metagenome-assembled genomes". Nature Microbiology. 7 (7): 962–973. doi:10.1038/s41564-022-01144-6. PMID 35760839.
- Tamarit D, Caceres EF, Krupovic M, Nijland R, Eme L, Robinson NP, Ettema TJ (July 2022). "A closed Candidatus Odinarchaeum chromosome exposes Asgard archaeal viruses". Nature Microbiology. 7 (7): 948–952. doi:10.1038/s41564-022-01122-y. PMC 9246712. PMID 35760836. S2CID 250090798.
- Rambo IM, Langwig MV, Leão P, De Anda V, Baker BJ (July 2022). "Genomes of six viruses that infect Asgard archaea from deep-sea sediments". Nature Microbiology. 7 (7): 953–961. doi:10.1038/s41564-022-01150-8. PMID 35760837.
- Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M (November 2017). "The enigmatic archaeal virosphere". Nature Reviews. Microbiology. 15 (12): 724–739. doi:10.1038/nrmicro.2017.125. PMID 29123227. S2CID 21789564.
- Alarcón-Schumacher T, Erdmann S (July 2022). "A trove of Asgard archaeal viruses". Nature Microbiology. 7 (7): 931–932. doi:10.1038/s41564-022-01148-2. PMID 35760838.
- Wu F, Speth DR, Philosof A, Crémière A, Narayanan A, Barco RA, et al. (February 2022). "Unique mobile elements and scalable gene flow at the prokaryote-eukaryote boundary revealed by circularized Asgard archaea genomes". Nature Microbiology. 7 (2): 200–212. doi:10.1038/s41564-021-01039-y. PMC 8813620. PMID 35027677.
External links
- Traci Watson: The trickster microbes that are shaking up the tree of life, in: Nature, 14 May 2019