Human milk microbiome
The human milk microbiota, also known as human milk probiotics (HMP), refers to the microbiota (community of microorganisms) residing in the human mammary glands and breast milk.[1] Human breast milk has been traditionally assumed to be sterile,[1][2] but more recently both microbial culture and culture-independent techniques have confirmed that human milk contains diverse communities of bacteria which are distinct from other microbial communities inhabiting the human body.[3][4][5]
The human milk microbiota which could be source of commensal, mutualistic, and potentially probiotic bacteria to the infant gut microbiota.[2] The World Health Organization (WHO) defines "probiotics" as "living organisms which when administered in adequate amounts confer a health benefit on the host".[6]
Occurrence
Breast milk is a natural source of lactic acid bacteria for the newborn through breastfeeding, and may be considered a symbiotic food.[7] The normal concentration of bacteria in milk from healthy women was about 103 colony-forming units (CFU) per milliliter.[8] The milk's bacterial communities were generally complex.[8] Among the hundreds of operational taxonomic units detected in the milk of every woman, only nine (Streptococcus, Staphylococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Propionibacterium, Sphingomonas, and Nitrobacteraceae) were present in every sample from every woman, but an individual's milk bacterial community was generally stable over time.[9] Human milk is a source of live Staphylococci, Streptococci, lactic acid bacteria, Bifidobacteria, Propionibacteria, Corynebacteria, and closely related Gram-positive bacteria for the infant gut.[2]
Composition
Breast milk was considered to be free of bacteria until about the early 2000s, when lactic acid bacteria were first described in human milk hygienically collected from healthy women.[7] Several studies have shown that there is a mother-to-infant transfer of bacterial strains belonging, at least, to the genera Lactobacillus, Staphylococcus, Enterococcus, and Bifidobacterium through breastfeeding, thus accounting for the close relationship of bacterial composition of the gut microbiota of breastfed infants with that found in the breast milk of their respective mothers.[2] Research has also found that there are similarities between human milk and infant gut microbial flora, suggesting that dietary exposure, such as human milk probiotics, may have a contribution in supporting infant gut microbiota and immune development.[10]
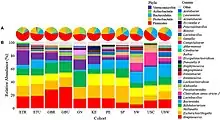
Bacteria commonly isolated in human milk samples include Bifidobacterium, Lactobacillus, Staphylococcus, Streptococcus, Bacteroides, Clostridium, Micrococcus, Enterococcus, and Escherichia.[3][5] Metagenomic analyses of human milk find it is dominated by Staphylococcus, Pseudomonas, and Edwardsiella.[11][12] The human milk microbiome likely varies by population and between individual women,[13] however, a study based on a group of U.S. women observed the same nine bacterial taxa in all samples from all of their participants, suggesting a common "core" of the milk microbiome, at least in that population.[8] Bacterial communities of human colostrum have been reported as being more diverse than those found in mature milk.[1][14]
The three strains of Lactobacilli with probiotic properties that were isolated from breast milk were L. fermentum CECT5716, L. gasseri CECT5714, and L. salivarius CECT5713,[15] with L. fermentum being one of the most abundant strains.[9] Early administration of L. fermentum CECT5716 in infant formula is claimed to be safe and well tolerated for infants one to six months of age,[16] and safe for long term use.[17]
Origin
While the origins of the human milk microbiome are not exactly known,[1] several hypotheses for its establishment have been proposed. Bacteria present in human milk may be derived from the surrounding breast skin flora,[18][19] or the infant's oral cavity microbiota.[8][12][20][21] Retrograde backflow during nursing or suckling may also lead to bacterial establishment in the mammary ducts,[22] supported by the observation that a certain degree of flowback has been shown to occur during nursing using infrared photography.[23] Alternatively, bacteria may be translocated to the mammary duct from the maternal gastrointestinal tract via an entero-mammary pathway, facilitated by dendritic cells.[2][3][24]
Environmental factors
Several factors may influence the composition of human milk probiotics, such as maternal body mass index (BMI), infant sex, birth modality, and mode of breastfeeding.[25][26] A study done by Soto et al also revealed that Lactobacilli and Bifidobacteria are more commonly found in the human milk of women who did not receive any antibiotics during pregnancy and lactation.[9]
Human milk oligosaccharides (HMOs), a primary component of human milk, are prebiotics which have been shown to promote growth of beneficial Bifidobacterium and Bacteroides species.[27][28][29]
Maternal health
Maternal health status is associated with changes in the bacterial composition of milk. Higher maternal BMI and obesity are associated with changes in the levels of Bifidobacterium and Staphylococcus species and overall lower bacterial diversity.[14][30] Milk of women with celiac disease is observed to have reduced levels of Bacteroides and Bifidobacterium.[31] Women who are HIV-positive show higher bacterial diversity and increased abundances of Lactobacillus in their milk than do non-HIV-positive women.[32] Mastitis has been linked to changes in human milk microbiota at the phylum level, lower microbial diversity, and decreased abundance of obligate anaerobic taxa.[33][34][12]
Women delivering term and preterm show differences in their milk microbiome composition, with mothers of term-births showing lower abundances of Enterococcus species and higher amounts of Bifidobacterium species in their milk compared to mothers of preterm births.[35]
Few studies have been conducted examining the influence of maternal diet on the milk microbiome,[1] but diet is known to influence other aspects of milk composition, such as the lipid profile,[36][37] which in turn could affect its microbial composition.[1] Variation in the fat and carbohydrate content of the maternal diet may influence the taxonomic composition of the milk microbiome.[38]
Both the taxonomic composition and diversity of bacteria present in human milk likely vary by maternal geographic location,[1][13][8] however studies with more geographically diverse participants are needed to better understand variation between populations.[1]
Maternal perinatal antibiotic use is associated with changes in the prevalence of Lactobacillus, Bifidobacterium, Staphylococcus, and Eubacterium in milk.[9][39][40]
Social network density of mother-infant dyads was found to be associated with increased bacterial diversity in the milk microbiome of mothers in the Central African Republic.[41]
Delivery method
Mode of delivery may influence composition of the human milk microbiome. Vaginal births is associated with high taxonomic diversity and high prevalence of Bifidobacterium and Lactobacillus, and the opposite trend being seen with birth by caesarean section,[14][35][42][9][43] however no relationship between delivery mode and the maternal milk microbiome has also been observed.[44]
Lactation stage
The human milk microbiome varies across lactation stage, with higher microbial diversity observed in colostrum than in mature milk.[1][14] Taxonomic composition of human milk also varies across the lactation period, initially dominated by Weissella, Leuconostoc, Staphylococcus, Streptococcus, and Lactococcus species,[14] and later composed primarily of Veillonella, Prevotella, Leptotrichia, Lactobacillus, Streptococcus, Bifidobacterium, and Enterococcus.[14][35]
Influences on health
Breastfeeding is thought to be an important driver of infant gut microbiome establishment.[45] The gut microbiome of breastfed infants is less diverse, contains higher amounts of Bifidobacterium and Lactobacillus species, and fewer potential pathogenic taxa than the gut microbiome of formula-fed infants.[46][47][48] Human milk bacteria may reduce risk of infection in breastfed infants by competitively excluding harmful bacteria,[49][50] and producing antimicrobial compounds which eliminate pathogenic strains.[51][52][53][49] Certain Lactobacilli and Bifidobacteria, the growth of which is stimulated by HMOs,[54] contribute to healthy metabolic and immune-related functioning in the infant gut.[55][56][2][57]
Benefits for breastfeeding mother
Research studies showed that L. fermentum could improve mastitis, a common inflammatory disease associated with lactation, by reducing the number of Streptococcus load which is believed to be the causal agent and risk factor of mastitis.[58]
Benefit for infants
Breastfed children have a lower incidence of infections than formula-fed children, which could be mediated in part through modulation of the intestinal microflora by breast milk components.[59] Indeed, breast-fed infants seem to develop a gut microflora richer in Lactobacilli and Bifidobacteria with reduced pathogenic bacteria compared with formula-fed infants.[60] Research, by Maldonado et al, found that infants receiving a follow-on formula enriched with L. fermentum demonstrated a reduction in gastrointestinal and respiratory infections, thus the administration of such formula may be useful for the prevention of community-acquired gastrointestinal and upper respiratory infections in infants.[16]
Human milk probiotics could also act as pioneering species to increase the colonization of ‘beneficial’ bacteria and support the infant’s immature immune system.[61] It is known that Lactobacilli and Bifidobacteria can suppress the growth of pathogenic microorganisms such as Salmonella typhimurium and Clostridium perfringens by colonization of a child's intestine and competing for nutrients, thus preventing their adhesion. Intestinal colonization by commensal bacteria also plays a vital role in maintaining homeostasis of immune system. These bacteria stimulate the T helper 1 response and counteract the trend towards a T helper 2 response of neonatal immune system, which in turn reducing the incidence of the inflammatory processes such as necrotizing enterocolitis.[15]
Children with colic symptoms possibly have an imbalance in the intestinal microbiota – analyses of faecal samples found higher counts of coliform bacteria and lower counts of Lactobacilli in infants with colic symptoms compared with children not suffering from colic.[62] On the other hand, probiotics have been shown to influence intestinal motility and sensory neurons as well as contractile activity of the intestine and to exert anti-inflammatory effects.[61]
Evolutionary implications
There is some indication of relationships between milk microbiota and other human milk components, including HMOs, maternal cells, and nutrient profiles.[29][63] Specific bacterial genera have been shown to be associated with variation in levels of milk macronutrients such as lactose, proteins, and fats.[63] HMOs selectively facilitate growth of particular beneficial bacteria, notably Bifidobacterium species.[64][65] Furthermore, as Bifidobacteria genomes are uniquely equipped to metabolize HMOs,[66] which are otherwise indigestible by enzymes of the infant gut, some have suggested a coevolution between HMOs and certain bacteria common in both the milk and infant gastrointestinal microbiomes.[67][68] Furthermore, relative to other mammalian milks such as primate milk, human milk appears to be unique with respect to the complexity and diversity of its oligosaccharide repertoire. Human milk is typified by greater overall HMO diversity and predominance of oligosaccharides known to promote growth of Bifidobacterium in the infant gut.[69] Milk microbiota are thought to play an essential role in programming the infant immune system, and tend to reduce the risk of adverse infant health outcomes.[57] Differences in milk oligosaccharides between humans and non-human primates could be indicative of variation in pathogen exposure associated with increased sociality and group sizes.[70] Together, these observations may indicate that milk microbial communities have coevolved with their human host,[68] supported by the expectation that microbes which promote host health facilitate their own transmission and proliferation.[71]
Comparisons with other mammals
Both human and macaque milks contains high abundances of Streptococcus and Lactobacillus bacteria, but differ in their respective relative abundances of these taxa.[72] Bacteria observed to be most common in healthy bovine milk include Ralstonia, Pseudomonas, Sphingomonas, Stenotrophomonas, Psychrobacter, Bradyrhizobium, Corynebacterium, Pelomonas, Staphylococcus, Faecalibacterium, Lachnospiraceae, Propionibacterium, Aeribacillus, Bacteroides, Streptococcus, Anaerococcus, Lactobacillus, Porphyromonas, Comamonas, Fusobacterium, and Enterococcus.[73][74][75][76]
See also
- Human microbiota
- Breast milk
- Human milk oligosaccharide
References
- Gomez-Gallego C, Garcia-Mantrana I, Salminen S, Collado MC (December 2016). "The human milk microbiome and factors influencing its composition and activity". Seminars in Fetal & Neonatal Medicine. 21 (6): 400–405. doi:10.1016/j.siny.2016.05.003. hdl:10261/162047. PMID 27286644.
- Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, Rodríguez JM (March 2013). "The human milk microbiota: origin and potential roles in health and disease". Pharmacological Research. 69 (1): 1–10. doi:10.1016/j.phrs.2012.09.001. PMID 22974824.
- Martín R, Jiménez E, Heilig H, Fernández L, Marín ML, Zoetendal EG, Rodríguez JM (February 2009). "Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR". Applied and Environmental Microbiology. 75 (4): 965–9. Bibcode:2009ApEnM..75..965M. doi:10.1128/aem.02063-08. PMC 2643565. PMID 19088308.
- Díaz-Ropero MP, Martín R, Sierra S, Lara-Villoslada F, Rodríguez JM, Xaus J, Olivares M (February 2007). "Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response". Journal of Applied Microbiology. 102 (2): 337–43. doi:10.1111/j.1365-2672.2006.03102.x. PMID 17241338.
- Collado MC, Delgado S, Maldonado A, Rodríguez JM (May 2009). "Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR". Letters in Applied Microbiology. 48 (5): 523–8. doi:10.1111/j.1472-765x.2009.02567.x. PMID 19228290.
- Food and Agriculture Organization and World Health Organization Expert Consultation. (2001). Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria (Report). Córdoba, Argentina: Food and Agriculture Organization of the United Nations and World Health Organization.
- Martín R, Langa S, Reviriego C, Jimínez E, Marín ML, Xaus J, et al. (December 2003). "Human milk is a source of lactic acid bacteria for the infant gut". The Journal of Pediatrics. 143 (6): 754–8. doi:10.1016/j.jpeds.2003.09.028. PMID 14657823.
- Hunt KM, Foster JA, Forney LJ, Schütte UM, Beck DL, Abdo Z, et al. (2011). "Characterization of the diversity and temporal stability of bacterial communities in human milk". PLOS ONE. 6 (6): e21313. Bibcode:2011PLoSO...621313H. doi:10.1371/journal.pone.0021313. PMC 3117882. PMID 21695057.
- Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L (July 2014). "Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors". Journal of Pediatric Gastroenterology and Nutrition. 59 (1): 78–88. doi:10.1097/MPG.0000000000000347. PMC 4086764. PMID 24590211.
- Martín V, Maldonado-Barragán A, Moles L, Rodriguez-Baños M, Campo RD, Fernández L, et al. (February 2012). "Sharing of bacterial strains between breast milk and infant feces". Journal of Human Lactation. 28 (1): 36–44. doi:10.1177/0890334411424729. PMID 22267318. S2CID 7489223.
- Ward TL, Hosid S, Ioshikhes I, Altosaar I (May 2013). "Human milk metagenome: a functional capacity analysis". BMC Microbiology. 13: 116. doi:10.1186/1471-2180-13-116. PMC 3679945. PMID 23705844.
- Jiménez E, de Andrés J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, et al. (August 2015). "Metagenomic Analysis of Milk of Healthy and Mastitis-Suffering Women". Journal of Human Lactation. 31 (3): 406–15. doi:10.1177/0890334415585078. PMID 25948578. S2CID 24138114.
- Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, et al. (2016). "Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles Across Specific Geographic Locations". Frontiers in Microbiology. 7: 1619. doi:10.3389/fmicb.2016.01619. PMC 5061857. PMID 27790209.
- Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A (September 2012). "The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery". The American Journal of Clinical Nutrition. 96 (3): 544–51. doi:10.3945/ajcn.112.037382. PMID 22836031.
- Lara-Villoslada F, Olivares M, Sierra S, Rodríguez JM, Boza J, Xaus J (October 2007). "Beneficial effects of probiotic bacteria isolated from breast milk". The British Journal of Nutrition. 98 Suppl 1: S96-100. doi:10.1017/S0007114507832910. PMID 17922969.
- Maldonado J, Cañabate F, Sempere L, Vela F, Sánchez AR, Narbona E, et al. (January 2012). "Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants". Journal of Pediatric Gastroenterology and Nutrition. 54 (1): 55–61. doi:10.1097/MPG.0b013e3182333f18. PMID 21873895. S2CID 29071981.
- Gil-Campos M, López MÁ, Rodriguez-Benítez MV, Romero J, Roncero I, Linares MD, et al. (February 2012). "Lactobacillus fermentum CECT 5716 is safe and well tolerated in infants of 1-6 months of age: a randomized controlled trial". Pharmacological Research. 65 (2): 231–8. doi:10.1016/j.phrs.2011.11.016. PMID 22155106.
- West PA, Hewitt JH, Murphy OM (April 1979). "Influence of methods of collection and storage on the bacteriology of human milk". The Journal of Applied Bacteriology. 46 (2): 269–77. doi:10.1111/j.1365-2672.1979.tb00820.x. PMID 572360.
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. (May 2009). "Topographical and temporal diversity of the human skin microbiome". Science. 324 (5931): 1190–2. Bibcode:2009Sci...324.1190G. doi:10.1126/science.1171700. PMC 2805064. PMID 19478181.
- Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, Meline BS, Swanson KS (August 2011). "Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing". PLOS ONE. 6 (8): e23503. Bibcode:2011PLoSO...623503C. doi:10.1371/journal.pone.0023503. PMC 3154475. PMID 21853142.
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M (April 2009). "Global diversity in the human salivary microbiome". Genome Research. 19 (4): 636–43. doi:10.1101/gr.084616.108. PMC 2665782. PMID 19251737.
- Rodríguez JM (November 2014). "The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation?". Advances in Nutrition. 5 (6): 779–84. doi:10.3945/an.114.007229. PMC 4224214. PMID 25398740.
- Ramsay DT, Kent JC, Owens RA, Hartmann PE (February 2004). "Ultrasound imaging of milk ejection in the breast of lactating women". Pediatrics. 113 (2): 361–7. doi:10.1542/peds.113.2.361. PMID 14754950.
- Jeurink PV, van Bergenhenegouwen J, Jiménez E, Knippels LM, Fernández L, Garssen J, et al. (March 2013). "Human milk: a source of more life than we imagine". Beneficial Microbes. 4 (1): 17–30. doi:10.3920/bm2012.0040. PMID 23271066.
- Munblit D, Peroni DG, Boix-Amorós A, Hsu PS, Van't Land B, Gay MC, et al. (August 2017). "Human Milk and Allergic Diseases: An Unsolved Puzzle". Nutrients. 9 (8): 894. doi:10.3390/nu9080894. PMC 5579687. PMID 28817095.
- Moossavi S, Khafipour E, Sepehri S, Robertson B, Bode L, Becker AB, et al. (2017). Maternal and early life factors influencing the human milk microbiota in the child cohort. Poster Session: Canadian Society of Microbiololists. Waterloo ON.
- Bode L (November 2009). "Human milk oligosaccharides: prebiotics and beyond". Nutrition Reviews. 67 Suppl 2 (suppl_2): S183-91. doi:10.1111/j.1753-4887.2009.00239.x. PMID 19906222.
- Jost T, Lacroix C, Braegger C, Chassard C (July 2015). "Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health". Nutrition Reviews. 73 (7): 426–37. doi:10.1093/nutrit/nuu016. PMID 26081453.
- Williams JE, Price WJ, Shafii B, Yahvah KM, Bode L, McGuire MA, McGuire MK (August 2017). "Relationships Among Microbial Communities, Maternal Cells, Oligosaccharides, and Macronutrients in Human Milk". Journal of Human Lactation. 33 (3): 540–551. doi:10.1177/0890334417709433. PMID 28609134.
- Collado MC, Laitinen K, Salminen S, Isolauri E (July 2012). "Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk". Pediatric Research. 72 (1): 77–85. doi:10.1038/pr.2012.42. PMID 22453296.
- Olivares M, Albrecht S, De Palma G, Ferrer MD, Castillejo G, Schols HA, Sanz Y (February 2015). "Human milk composition differs in healthy mothers and mothers with celiac disease". European Journal of Nutrition. 54 (1): 119–28. doi:10.1007/s00394-014-0692-1. PMID 24700375. S2CID 36508893.
- González R, Maldonado A, Martín V, Mandomando I, Fumadó V, Metzner KJ, et al. (November 2013). "Breast milk and gut microbiota in African mothers and infants from an area of high HIV prevalence". PLOS ONE. 8 (11): e80299. Bibcode:2013PLoSO...880299G. doi:10.1371/journal.pone.0080299. PMC 3841168. PMID 24303004.
- Patel SH, Vaidya YH, Patel RJ, Pandit RJ, Joshi CG, Kunjadiya AP (August 2017). "Culture independent assessment of human milk microbial community in lactational mastitis". Scientific Reports. 7 (1): 7804. Bibcode:2017NatSR...7.7804P. doi:10.1038/s41598-017-08451-7. PMC 5552812. PMID 28798374.
- Delgado S, Arroyo R, Martín R, Rodríguez JM (April 2008). "PCR-DGGE assessment of the bacterial diversity of breast milk in women with lactational infectious mastitis". BMC Infectious Diseases. 8: 51. doi:10.1186/1471-2334-8-51. PMC 2383900. PMID 18423017.
- Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C (August 2014). "Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota". Journal of Perinatology. 34 (8): 599–605. doi:10.1038/jp.2014.47. PMID 24674981. S2CID 37695304.
- Nishimura RY, Barbieiri P, Castro GS, Jordão AA, Perdoná G, Sartorelli DS (June 2014). "Dietary polyunsaturated fatty acid intake during late pregnancy affects fatty acid composition of mature breast milk". Nutrition. 30 (6): 685–9. doi:10.1016/j.nut.2013.11.002. PMID 24613435.
- Peng Y, Zhou T, Wang Q, Liu P, Zhang T, Zetterström R, Strandvik B (2009). "Fatty acid composition of diet, cord blood and breast milk in Chinese mothers with different dietary habits". Prostaglandins, Leukotrienes, and Essential Fatty Acids. 81 (5–6): 325–30. doi:10.1016/j.plefa.2009.07.004. PMID 19709866.
- Meyer KM, Mohammad M, Bode L, Chu DM, Ma J, Haymond M, Aagaard K (2017). "20: Maternal diet structures the breast milk microbiome in association with human milk oligosaccharides and gut-associated bacteria". American Journal of Obstetrics and Gynecology. 216 (1): S15. doi:10.1016/j.ajog.2016.11.911.
- Witt A, Mason MJ, Burgess K, Flocke S, Zyzanski S (January 2014). "A case control study of bacterial species and colony count in milk of breastfeeding women with chronic pain". Breastfeeding Medicine. 9 (1): 29–34. doi:10.1089/bfm.2013.0012. PMC 3903327. PMID 23789831.
- Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. (May 2014). "Microbiota of human breast tissue". Applied and Environmental Microbiology. 80 (10): 3007–14. Bibcode:2014ApEnM..80.3007U. doi:10.1128/aem.00242-14. PMC 4018903. PMID 24610844.
- Meehan CL, Lackey KA, Hagen EH, Williams JE, Roulette J, Helfrecht C, et al. (July 2018). "Social networks, cooperative breeding, and the human milk microbiome". American Journal of Human Biology. 30 (4): e23131. doi:10.1002/ajhb.23131. PMID 29700885. S2CID 13793532.
- Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC (February 2016). "Impact of mode of delivery on the milk microbiota composition of healthy women". Journal of Developmental Origins of Health and Disease. 7 (1): 54–60. doi:10.1017/S2040174415001397. PMID 26286040. S2CID 10368527.
- Hoashi M, Meche L, Mahal LK, Bakacs E, Nardella D, Naftolin F, et al. (July 2016). "Human Milk Bacterial and Glycosylation Patterns Differ by Delivery Mode". Reproductive Sciences. 23 (7): 902–7. doi:10.1177/1933719115623645. PMID 26711314. S2CID 38606958.
- Urbaniak C, Angelini M, Gloor GB, Reid G (January 2016). "Human milk microbiota profiles in relation to birthing method, gestation and infant gender". Microbiome. 4 (1): 1. doi:10.1186/s40168-015-0145-y. PMC 4702315. PMID 26739322.
- Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. (December 2017). "The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota". Microbiology and Molecular Biology Reviews. 81 (4): e00036–17. doi:10.1128/mmbr.00036-17. PMC 5706746. PMID 29118049.
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. (May 2012). "Human gut microbiome viewed across age and geography". Nature. 486 (7402): 222–7. Bibcode:2012Natur.486..222Y. doi:10.1038/nature11053. PMC 3376388. PMID 22699611.
- O'Sullivan A, Farver M, Smilowitz JT (2015). "The Influence of Early Infant-Feeding Practices on the Intestinal Microbiome and Body Composition in Infants". Nutrition and Metabolic Insights. 8 (Suppl 1): 1–9. doi:10.4137/NMI.S29530. PMC 4686345. PMID 26715853.
- Bezirtzoglou E, Tsiotsias A, Welling GW (December 2011). "Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH)". Anaerobe. 17 (6): 478–82. doi:10.1016/j.anaerobe.2011.03.009. PMID 21497661.
- Olivares M, Díaz-Ropero MP, Martín R, Rodríguez JM, Xaus J (July 2006). "Antimicrobial potential of four Lactobacillus strains isolated from breast milk". Journal of Applied Microbiology. 101 (1): 72–9. doi:10.1111/j.1365-2672.2006.02981.x. PMID 16834593.
- Heikkilä MP, Saris PE (2003). "Inhibition of Staphylococcus aureus by the commensal bacteria of human milk". Journal of Applied Microbiology. 95 (3): 471–8. doi:10.1046/j.1365-2672.2003.02002.x. PMID 12911694.
- Beasley SS, Saris PE (August 2004). "Nisin-producing Lactococcus lactis strains isolated from human milk". Applied and Environmental Microbiology. 70 (8): 5051–3. Bibcode:2004ApEnM..70.5051B. doi:10.1128/aem.70.8.5051-5053.2004. PMC 492443. PMID 15294850.
- Martín R, Olivares M, Marín ML, Fernández L, Xaus J, Rodríguez JM (February 2005). "Probiotic potential of 3 Lactobacilli strains isolated from breast milk". Journal of Human Lactation. 21 (1): 8–17, quiz 18-21, 41. doi:10.1177/0890334404272393. PMID 15681631. S2CID 23135001.
- Martín R, Jiménez E, Olivares M, Marín ML, Fernández L, Xaus J, Rodríguez JM (October 2006). "Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair". International Journal of Food Microbiology. 112 (1): 35–43. doi:10.1016/j.ijfoodmicro.2006.06.011. PMID 16843562.
- Bode L (September 2012). "Human milk oligosaccharides: every baby needs a sugar mama". Glycobiology. 22 (9): 1147–62. doi:10.1093/glycob/cws074. PMC 3406618. PMID 22513036.
- Zivkovic AM, German JB, Lebrilla CB, Mills DA (March 2011). "Human milk glycobiome and its impact on the infant gastrointestinal microbiota". Proceedings of the National Academy of Sciences of the United States of America. 108 Suppl 1 (Supplement 1): 4653–8. Bibcode:2011PNAS..108.4653Z. doi:10.1073/pnas.1000083107. PMC 3063602. PMID 20679197.
- Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, et al. (October 2011). "Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria". The Journal of Biological Chemistry. 286 (40): 34583–92. doi:10.1074/jbc.M111.248138. PMC 3186357. PMID 21832085.
- Donnet-Hughes A, Perez PF, Doré J, Leclerc M, Levenez F, Benyacoub J, et al. (August 2010). "Potential role of the intestinal microbiota of the mother in neonatal immune education". The Proceedings of the Nutrition Society. 69 (3): 407–15. doi:10.1017/S0029665110001898. PMID 20633308.
- Arroyo R, Martín V, Maldonado A, Jiménez E, Fernández L, Rodríguez JM (June 2010). "Treatment of infectious mastitis during lactation: antibiotics versus oral administration of Lactobacilli isolated from breast milk". Clinical Infectious Diseases. 50 (12): 1551–8. doi:10.1086/652763. PMID 20455694.
- Wold AE, Adlerberth I (2000). "Breast feeding and the intestinal microflora of the infant--implications for protection against infectious diseases". Advances in Experimental Medicine and Biology. 478: 77–93. doi:10.1007/0-306-46830-1_7. ISBN 0-306-46405-5. PMID 11065062.
- Mackie RI, Sghir A, Gaskins HR (May 1999). "Developmental microbial ecology of the neonatal gastrointestinal tract". The American Journal of Clinical Nutrition. 69 (5): 1035S–1045S. doi:10.1093/ajcn/69.5.1035s. PMID 10232646.
- Bergmann H, Rodríguez JM, Salminen S, Szajewska H (October 2014). "Probiotics in human milk and probiotic supplementation in infant nutrition: a workshop report". The British Journal of Nutrition. 112 (7): 1119–28. doi:10.1017/S0007114514001949. PMID 25160058.
- Savino F, Tarasco V (December 2010). "New treatments for infant colic". Current Opinion in Pediatrics. 22 (6): 791–7. doi:10.1097/MOP.0b013e32833fac24. PMID 20859207. S2CID 26003983.
- Boix-Amorós A, Collado MC, Mira A (2016). "Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation". Frontiers in Microbiology. 7: 492. doi:10.3389/fmicb.2016.00492. PMC 4837678. PMID 27148183.
- Clancy KB, Hinde K, Rutherford JN (2013). Building babies: primate development in proximate and ultimate perspective. Developments in Primatology: Progress and Prospects. New York: Springer. doi:10.1007/978-1-4614-4060-4_11. ISBN 978-1-4614-4059-8.
- Sela DA, Mills DA (July 2010). "Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides". Trends in Microbiology. 18 (7): 298–307. doi:10.1016/j.tim.2010.03.008. PMC 2902656. PMID 20409714.
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, et al. (December 2008). "The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome". Proceedings of the National Academy of Sciences of the United States of America. 105 (48): 18964–9. Bibcode:2008PNAS..10518964S. doi:10.1073/pnas.0809584105. PMC 2596198. PMID 19033196.
- German JB, Freeman SL, Lebrilla CB, Mills DA (2008). "Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria". Nestle Nutrition Workshop Series. Paediatric Programme. Nestlé Nutrition Workshop Series: Pediatric Program. 62: 205–18, discussion 218-22. doi:10.1159/000146322. ISBN 978-3-8055-8553-8. PMC 2861563. PMID 18626202.
- Allen-Blevins CR, Sela DA, Hinde K (April 2015). "Milk bioactives may manipulate microbes to mediate parent-offspring conflict". Evolution, Medicine, and Public Health. 2015 (1): 106–21. doi:10.1093/emph/eov007. PMC 4512713. PMID 25835022.
- Urashima T, Odaka G, Asakuma S, Uemura Y, Goto K, Senda A, et al. (May 2009). "Chemical characterization of oligosaccharides in chimpanzee, bonobo, gorilla, orangutan, and siamang milk or colostrum". Glycobiology. 19 (5): 499–508. doi:10.1093/glycob/cwp006. PMID 19164487.
- Tao N, Wu S, Kim J, An HJ, Hinde K, Power ML, et al. (April 2011). "Evolutionary glycomics: characterization of milk oligosaccharides in primates". Journal of Proteome Research. 10 (4): 1548–57. doi:10.1021/pr1009367. PMC 3070053. PMID 21214271.
- Funkhouser LJ, Bordenstein SR (2013). "Mom knows best: the universality of maternal microbial transmission". PLOS Biology. 11 (8): e1001631. doi:10.1371/journal.pbio.1001631. PMC 3747981. PMID 23976878.
- Jin L, Hinde K, Tao L (February 2011). "Species diversity and relative abundance of lactic acid bacteria in the milk of rhesus monkeys (Macaca mulatta)". Journal of Medical Primatology. 40 (1): 52–8. doi:10.1111/j.1600-0684.2010.00450.x. PMC 3697067. PMID 20946146.
- Kuehn JS, Gorden PJ, Munro D, Rong R, Dong Q, Plummer PJ, et al. (April 2013). "Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis". PLOS ONE. 8 (4): e61959. Bibcode:2013PLoSO...861959K. doi:10.1371/journal.pone.0061959. PMC 3636265. PMID 23634219.
- Oikonomou G, Bicalho ML, Meira E, Rossi RE, Foditsch C, Machado VS, et al. (2014). "Microbiota of cow's milk; distinguishing healthy, sub-clinically and clinically diseased quarters". PLOS ONE. 9 (1): e85904. Bibcode:2014PLoSO...985904O. doi:10.1371/journal.pone.0085904. PMC 3896433. PMID 24465777.
- Zhang R, Huo W, Zhu W, Mao S (March 2015). "Characterization of bacterial community of raw milk from dairy cows during subacute ruminal acidosis challenge by high-throughput sequencing". Journal of the Science of Food and Agriculture. 95 (5): 1072–9. doi:10.1002/jsfa.6800. PMID 24961605.
- Addis MF, Tanca A, Uzzau S, Oikonomou G, Bicalho RC, Moroni P (July 2016). "The bovine milk microbiota: insights and perspectives from -omics studies". Molecular BioSystems. 12 (8): 2359–72. doi:10.1039/c6mb00217j. hdl:2434/383895. PMID 27216801.