Introduction to viruses
A virus is a tiny infectious agent that reproduces inside the cells of living hosts. When infected, the host cell is forced to rapidly produce thousands of identical copies of the original virus. Unlike most living things, viruses do not have cells that divide; new viruses assemble in the infected host cell. But unlike simpler infectious agents like prions, they contain genes, which allow them to mutate and evolve. Over 4,800 species of viruses have been described in detail[1] out of the millions in the environment. Their origin is unclear: some may have evolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria.

Viruses are made of either two or three parts. All include genes. These genes contain the encoded biological information of the virus and are built from either DNA or RNA. All viruses are also covered with a protein coat to protect the genes. Some viruses may also have an envelope of fat-like substance that covers the protein coat, and makes them vulnerable to soap. A virus with this "viral envelope" uses it—along with specific receptors—to enter a new host cell. Viruses vary in shape from the simple helical and icosahedral to more complex structures. Viruses range in size from 20 to 300 nanometres; it would take 33,000 to 500,000 of them, side by side, to stretch to 1 centimetre (0.4 in).
Viruses spread in many ways. Although many are very specific about which host species or tissue they attack, each species of virus relies on a particular method to copy itself. Plant viruses are often spread from plant to plant by insects and other organisms, known as vectors. Some viruses of humans and other animals are spread by exposure to infected bodily fluids. Viruses such as influenza are spread through the air by droplets of moisture when people cough or sneeze. Viruses such as norovirus are transmitted by the faecal–oral route, which involves the contamination of hands, food and water. Rotavirus is often spread by direct contact with infected children. The human immunodeficiency virus, HIV, is transmitted by bodily fluids transferred during sex. Others, such as the dengue virus, are spread by blood-sucking insects.
Viruses, especially those made of RNA, can mutate rapidly to give rise to new types. Hosts may have little protection against such new forms. Influenza virus, for example, changes often, so a new vaccine is needed each year. Major changes can cause pandemics, as in the 2009 swine influenza that spread to most countries. Often, these mutations take place when the virus has first infected other animal hosts. Some examples of such "zoonotic" diseases include coronavirus in bats, and influenza in pigs and birds, before those viruses were transferred to humans.
Viral infections can cause disease in humans, animals and plants. In healthy humans and animals, infections are usually eliminated by the immune system, which can provide lifetime immunity to the host for that virus. Antibiotics, which work against bacteria, have no impact, but antiviral drugs can treat life-threatening infections. Those vaccines that produce lifelong immunity can prevent some infections.
Discovery

In 1884, French microbiologist Charles Chamberland invented the Chamberland filter (or Chamberland–Pasteur filter), that contains pores smaller than bacteria. He could then pass a solution containing bacteria through the filter, and completely remove them. In the early 1890s, Russian biologist Dmitri Ivanovsky used this method to study what became known as the tobacco mosaic virus. His experiments showed that extracts from the crushed leaves of infected tobacco plants remain infectious after filtration.[2]
At the same time, several other scientists showed that, although these agents (later called viruses) were different from bacteria and about one hundred times smaller, they could still cause disease. In 1899, Dutch microbiologist Martinus Beijerinck observed that the agent only multiplied when in dividing cells. He called it a "contagious living fluid" (Latin: contagium vivum fluidum)—or a "soluble living germ" because he could not find any germ-like particles.[3] In the early 20th century, English bacteriologist Frederick Twort discovered viruses that infect bacteria,[4] and French-Canadian microbiologist Félix d'Herelle described viruses that, when added to bacteria growing on agar, would lead to the formation of whole areas of dead bacteria. Counting these dead areas allowed him to calculate the number of viruses in the suspension.[5]
The invention of the electron microscope in 1931 brought the first images of viruses.[6] In 1935, American biochemist and virologist Wendell Meredith Stanley examined the tobacco mosaic virus (TMV) and found it to be mainly made from protein.[7] A short time later, this virus was shown to be made from protein and RNA.[8] Rosalind Franklin developed X-ray crystallographic pictures and determined the full structure of TMV in 1955.[9] Franklin confirmed that viral proteins formed a spiral hollow tube, wrapped by RNA, and also showed that viral RNA was a single strand, not a double helix like DNA.[10]
A problem for early scientists was that they did not know how to grow viruses without using live animals. The breakthrough came in 1931, when American pathologists Ernest William Goodpasture and Alice Miles Woodruff grew influenza, and several other viruses, in fertilised chickens' eggs.[11] Some viruses could not be grown in chickens' eggs. This problem was solved in 1949, when John Franklin Enders, Thomas Huckle Weller, and Frederick Chapman Robbins grew polio virus in cultures of living animal cells.[12] Over 4,800 species of viruses have been described in detail.[1]
Origins
Viruses co-exist with life wherever it occurs. They have probably existed since living cells first evolved. Their origin remains unclear because they do not fossilize, so molecular techniques have been the best way to hypothesise about how they arose. These techniques rely on the availability of ancient viral DNA or RNA, but most viruses that have been preserved and stored in laboratories are less than 90 years old.[13][14] Molecular methods have only been successful in tracing the ancestry of viruses that evolved in the 20th century.[15] New groups of viruses might have repeatedly emerged at all stages of the evolution of life.[16] There are three major theories about the origins of viruses:[16][17]
- Regressive theory
- Viruses may have once been small cells that parasitised larger cells. Eventually, the genes they no longer needed for a parasitic way of life were lost. The bacteria Rickettsia and Chlamydia are living cells that, like viruses, can reproduce only inside host cells. This lends credence to this theory, as their dependence on being parasites may have led to the loss of the genes that once allowed them to live on their own.[18]
- Cellular origin theory
- Some viruses may have evolved from bits of DNA or RNA that "escaped" from the genes of a larger organism. The escaped DNA could have come from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria.[19]
- Coevolution theory
- Viruses may have evolved from complex molecules of protein and DNA at the same time as cells first appeared on earth, and would have depended on cellular life for many millions of years.[20]
There are problems with all of these theories. The regressive hypothesis does not explain why even the smallest of cellular parasites do not resemble viruses in any way. The escape or the cellular origin hypothesis does not explain the presence of unique structures in viruses that do not appear in cells. The coevolution, or "virus-first" hypothesis, conflicts with the definition of viruses, because viruses depend on host cells.[20][21] Also, viruses are recognised as ancient, and to have origins that pre-date the divergence of life into the three domains.[22] This discovery has led modern virologists to reconsider and re-evaluate these three classical hypotheses.[16][22]
Structure
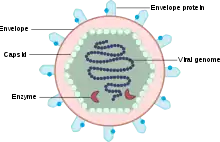
A virus particle, also called a virion, consists of genes made from DNA or RNA which are surrounded by a protective coat of protein called a capsid.[23] The capsid is made of many smaller, identical protein molecules called capsomers. The arrangement of the capsomers can either be icosahedral (20-sided), helical, or more complex. There is an inner shell around the DNA or RNA called the nucleocapsid, made out of proteins. Some viruses are surrounded by a bubble of lipid (fat) called an envelope, which makes them vulnerable to soap and alcohol.[24]
Size

Viruses are among the smallest infectious agents, and are too small to be seen by light microscopy; most of them can only be seen by electron microscopy. Their sizes range from 20 to 300 nanometres; it would take 30,000 to 500,000 of them, side by side, to stretch to one centimetre (0.4 in).[23] In comparison, bacteria are typically around 1000 nanometres (1 micrometer) in diameter, and host cells of higher organisms are typically a few tens of micrometers. Some viruses such as megaviruses and pandoraviruses are relatively large viruses. At around 1000 nanometres, these viruses, which infect amoebae, were discovered in 2003 and 2013.[25][26] They are around ten times wider (and thus a thousand times larger in volume) than influenza viruses, and the discovery of these "giant" viruses astonished scientists.[27]
Genes
The genes of viruses are made from DNA (deoxyribonucleic acid) and, in many viruses, RNA (ribonucleic acid). The biological information contained in an organism is encoded in its DNA or RNA. Most organisms use DNA, but many viruses have RNA as their genetic material. The DNA or RNA of viruses consists of either a single strand or a double helix.[28]
Viruses can reproduce rapidly because they have relatively few genes. For example, influenza virus has only eight genes and rotavirus has eleven. In comparison, humans have 20,000–25,000. Some viral genes contain the code to make the structural proteins that form the virus particle. Other genes make non-structural proteins found only in the cells the virus infects.[29][30]
All cells, and many viruses, produce proteins that are enzymes that drive chemical reactions. Some of these enzymes, called DNA polymerase and RNA polymerase, make new copies of DNA and RNA. A virus's polymerase enzymes are often much more efficient at making DNA and RNA than the equivalent enzymes of the host cells,[31] but viral RNA polymerase enzymes are error-prone, causing RNA viruses to mutate and form new strains.[32]
In some species of RNA virus, the genes are not on a continuous molecule of RNA, but are separated. The influenza virus, for example, has eight separate genes made of RNA. When two different strains of influenza virus infect the same cell, these genes can mix and produce new strains of the virus in a process called reassortment.[33]
Protein synthesis
Proteins are essential to life. Cells produce new protein molecules from amino acid building blocks based on information coded in DNA. Each type of protein is a specialist that usually only performs one function, so if a cell needs to do something new, it must make a new protein. Viruses force the cell to make new proteins that the cell does not need, but are needed for the virus to reproduce. Protein synthesis consists of two major steps: transcription and translation.[34]
Transcription is the process where information in DNA, called the genetic code, is used to produce RNA copies called messenger RNA (mRNA). These migrate through the cell and carry the code to ribosomes where it is used to make proteins. This is called translation because the protein's amino acid structure is determined by the mRNA's code. Information is hence translated from the language of nucleic acids to the language of amino acids.[34]
Some nucleic acids of RNA viruses function directly as mRNA without further modification. For this reason, these viruses are called positive-sense RNA viruses.[35] In other RNA viruses, the RNA is a complementary copy of mRNA and these viruses rely on the cell's or their own enzyme to make mRNA. These are called negative-sense RNA viruses. In viruses made from DNA, the method of mRNA production is similar to that of the cell. The species of viruses called retroviruses behave completely differently: they have RNA, but inside the host cell a DNA copy of their RNA is made with the help of the enzyme reverse transcriptase. This DNA is then incorporated into the host's own DNA, and copied into mRNA by the cell's normal pathways.[36]
Life-cycle
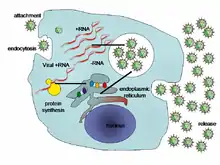
When a virus infects a cell, the virus forces it to make thousands more viruses. It does this by making the cell copy the virus's DNA or RNA, making viral proteins, which all assemble to form new virus particles.[37]
There are six basic, overlapping stages in the life cycle of viruses in living cells:[38]
- Attachment is the binding of the virus to specific molecules on the surface of the cell. This specificity restricts the virus to a very limited type of cell. For example, the human immunodeficiency virus (HIV) infects only human T cells, because its surface protein, gp120, can only react with CD4 and other molecules on the T cell's surface. Plant viruses can only attach to plant cells and cannot infect animals. This mechanism has evolved to favour those viruses that only infect cells in which they are capable of reproducing.
- Penetration follows attachment; viruses penetrate the host cell by endocytosis or by fusion with the cell.
- Uncoating happens inside the cell when the viral capsid is removed and destroyed by viral enzymes or host enzymes, thereby exposing the viral nucleic acid.
- Replication of virus particles is the stage where a cell uses viral messenger RNA in its protein synthesis systems to produce viral proteins. The RNA or DNA synthesis abilities of the cell produce the virus's DNA or RNA.
- Assembly takes place in the cell when the newly created viral proteins and nucleic acid combine to form hundreds of new virus particles.
- Release occurs when the new viruses escape or are released from the cell. Most viruses achieve this by making the cells burst, a process called lysis. Other viruses such as HIV are released more gently by a process called budding.
Effects on the host cell
Viruses have an extensive range of structural and biochemical effects on the host cell.[39]These are called cytopathic effects.[40] Most virus infections eventually result in the death of the host cell. The causes of death include cell lysis (bursting), alterations to the cell's surface membrane and apoptosis (cell "suicide").[41] Often cell death is caused by cessation of its normal activity due to proteins produced by the virus, not all of which are components of the virus particle.[42]
Some viruses cause no apparent changes to the infected cell. Cells in which the virus is latent (inactive) show few signs of infection and often function normally.[43] This causes persistent infections and the virus is often dormant for many months or years. This is often the case with herpes viruses.[44][45]
Some viruses, such as Epstein–Barr virus, often cause cells to proliferate without causing malignancy;[46] but some other viruses, such as papillomavirus, are an established cause of cancer.[47] When a cell's DNA is damaged by a virus such that the cell cannot repair itself, this often triggers apoptosis. One of the results of apoptosis is destruction of the damaged DNA by the cell itself. Some viruses have mechanisms to limit apoptosis so that the host cell does not die before progeny viruses have been produced; HIV, for example, does this.[48]
Viruses and diseases
There are many ways in which viruses spread from host to host but each species of virus uses only one or two. Many viruses that infect plants are carried by organisms; such organisms are called vectors. Some viruses that infect animals, including humans, are also spread by vectors, usually blood-sucking insects, but direct transmission is more common. Some virus infections, such as norovirus and rotavirus, are spread by contaminated food and water, by hands and communal objects, and by intimate contact with another infected person, while others are airborne (influenza virus). Viruses such as HIV, hepatitis B and hepatitis C are often transmitted by unprotected sex or contaminated hypodermic needles. To prevent infections and epidemics, it is important to know how each different kind of virus is spread.[49]
In humans
Common human diseases caused by viruses include the common cold, influenza, chickenpox and cold sores. Serious diseases such as Ebola and AIDS are also caused by viruses.[50] Many viruses cause little or no disease and are said to be "benign". The more harmful viruses are described as virulent.[51] Viruses cause different diseases depending on the types of cell that they infect. Some viruses can cause lifelong or chronic infections where the viruses continue to reproduce in the body despite the host's defence mechanisms.[52] This is common in hepatitis B virus and hepatitis C virus infections. People chronically infected with a virus are known as carriers. They serve as important reservoirs of the virus.[53][54]
Endemic
If the proportion of carriers in a given population reaches a given threshold, a disease is said to be endemic.[55] Before the advent of vaccination, infections with viruses were common and outbreaks occurred regularly. In countries with a temperate climate, viral diseases are usually seasonal. Poliomyelitis, caused by poliovirus often occurred in the summer months.[56] By contrast colds, influenza and rotavirus infections are usually a problem during the winter months.[57][58] Other viruses, such as measles virus, caused outbreaks regularly every third year.[59] In developing countries, viruses that cause respiratory and enteric infections are common throughout the year. Viruses carried by insects are a common cause of diseases in these settings. Zika and dengue viruses for example are transmitted by the female Aedes mosquitoes, which bite humans particularly during the mosquitoes' breeding season.[60]
Pandemic and emergent

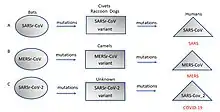
Although viral pandemics are rare events, HIV—which evolved from viruses found in monkeys and chimpanzees—has been pandemic since at least the 1980s.[62] During the 20th century there were four pandemics caused by influenza virus and those that occurred in 1918, 1957 and 1968 were severe.[63] Before its eradication, smallpox was a cause of pandemics for more than 3,000 years.[64] Throughout history, human migration has aided the spread of pandemic infections; first by sea and in modern times also by air.[65]
With the exception of smallpox, most pandemics are caused by newly evolved viruses. These "emergent" viruses are usually mutants of less harmful viruses that have circulated previously either in humans or in other animals.[66]
Severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) are caused by new types of coronaviruses. Other coronaviruses are known to cause mild infections in humans,[67] so the virulence and rapid spread of SARS infections—that by July 2003 had caused around 8,000 cases and 800 deaths—was unexpected and most countries were not prepared.[68]
A related coronavirus emerged in Wuhan, China, in November 2019 and spread rapidly around the world. Thought to have originated in bats and subsequently named severe acute respiratory syndrome coronavirus 2, infections with the virus cause a disease called COVID-19, that varies in severity from mild to deadly,[69] and led to a pandemic in 2020.[61][70][71] Restrictions unprecedented in peacetime were placed on international travel,[72] and curfews imposed in several major cities worldwide.[73]
In plants

There are many types of plant virus, but often they only cause a decrease in yield, and it is not economically viable to try to control them. Plant viruses are frequently spread from plant to plant by organisms called "vectors". These are normally insects, but some fungi, nematode worms and single-celled organisms have also been shown to be vectors. When control of plant virus infections is considered economical (perennial fruits, for example) efforts are concentrated on killing the vectors and removing alternate hosts such as weeds.[74] Plant viruses are harmless to humans and other animals because they can only reproduce in living plant cells.[75]
Bacteriophages
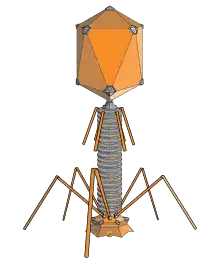
Bacteriophages are viruses that infect bacteria and archaea.[76] They are important in marine ecology: as the infected bacteria burst, carbon compounds are released back into the environment, which stimulates fresh organic growth. Bacteriophages are useful in scientific research because they are harmless to humans and can be studied easily. These viruses can be a problem in industries that produce food and drugs by fermentation and depend on healthy bacteria. Some bacterial infections are becoming difficult to control with antibiotics, so there is a growing interest in the use of bacteriophages to treat infections in humans.[77]
Innate immunity of animals
Animals, including humans, have many natural defences against viruses. Some are non-specific and protect against many viruses regardless of the type. This innate immunity is not improved by repeated exposure to viruses and does not retain a "memory" of the infection. The skin of animals, particularly its surface, which is made from dead cells, prevents many types of viruses from infecting the host. The acidity of the contents of the stomach destroys many viruses that have been swallowed. When a virus overcomes these barriers and enters the host, other innate defences prevent the spread of infection in the body. A special hormone called interferon is produced by the body when viruses are present, and this stops the viruses from reproducing by killing the infected cells and their close neighbours. Inside cells, there are enzymes that destroy the RNA of viruses. This is called RNA interference. Some blood cells engulf and destroy other virus-infected cells.[78]
Adaptive immunity of animals
Specific immunity to viruses develops over time and white blood cells called lymphocytes play a central role. Lymphocytes retain a "memory" of virus infections and produce many special molecules called antibodies. These antibodies attach to viruses and stop the virus from infecting cells. Antibodies are highly selective and attack only one type of virus. The body makes many different antibodies, especially during the initial infection. After the infection subsides, some antibodies remain and continue to be produced, usually giving the host lifelong immunity to the virus.[79]
Plant resistance
Plants have elaborate and effective defence mechanisms against viruses. One of the most effective is the presence of so-called resistance (R) genes. Each R gene confers resistance to a particular virus by triggering localised areas of cell death around the infected cell, which can often be seen with the unaided eye as large spots. This stops the infection from spreading.[80] RNA interference is also an effective defence in plants.[81] When they are infected, plants often produce natural disinfectants that destroy viruses, such as salicylic acid, nitric oxide and reactive oxygen molecules.[82]
Resistance to bacteriophages
The major way bacteria defend themselves from bacteriophages is by producing enzymes which destroy foreign DNA. These enzymes, called restriction endonucleases, cut up the viral DNA that bacteriophages inject into bacterial cells.[83]
Vaccines

Vaccines simulate a natural infection and its associated immune response, but do not cause the disease. Their use has resulted in the eradication of smallpox and a dramatic decline in illness and death caused by infections such as polio, measles, mumps and rubella.[84] Vaccines are available to prevent over fourteen viral infections of humans[85] and more are used to prevent viral infections of animals.[86] Vaccines may consist of either live or killed viruses.[87] Live vaccines contain weakened forms of the virus, but these vaccines can be dangerous when given to people with weak immunity. In these people, the weakened virus can cause the original disease.[88] Biotechnology and genetic engineering techniques are used to produce "designer" vaccines that only have the capsid proteins of the virus. Hepatitis B vaccine is an example of this type of vaccine.[89] These vaccines are safer because they can never cause the disease.[87]
Antiviral drugs

Since the mid-1980s, the development of antiviral drugs has increased rapidly, mainly driven by the AIDS pandemic. Antiviral drugs are often nucleoside analogues, which masquerade as DNA building blocks (nucleosides). When the replication of virus DNA begins, some of the fake building blocks are used. This prevents DNA replication because the drugs lack the essential features that allow the formation of a DNA chain. When DNA production stops the virus can no longer reproduce.[90] Examples of nucleoside analogues are aciclovir for herpes virus infections and lamivudine for HIV and hepatitis B virus infections. Aciclovir is one of the oldest and most frequently prescribed antiviral drugs.[91]
Other antiviral drugs target different stages of the viral life cycle. HIV is dependent on an enzyme called the HIV-1 protease for the virus to become infectious. There is a class of drugs called protease inhibitors, which bind to this enzyme and stop it from functioning.[92]
Hepatitis C is caused by an RNA virus. In 80% of those infected, the disease becomes chronic, and they remain infectious for the rest of their lives unless they are treated. There are effective treatments that use direct-acting antivirals.[93] Treatments for chronic carriers of the hepatitis B virus have been developed by a similar strategy, using lamivudine and other anti-viral drugs. In both diseases, the drugs stop the virus from reproducing and the interferon kills any remaining infected cells.[94]
HIV infections are usually treated with a combination of antiviral drugs, each targeting a different stage in the virus's life cycle. There are drugs that prevent the virus from attaching to cells, others that are nucleoside analogues and some poison the virus's enzymes that it needs to reproduce. The success of these drugs is proof of the importance of knowing how viruses reproduce.[92]
Role in ecology
Viruses are the most abundant biological entity in aquatic environments;[95] one teaspoon of seawater contains about ten million viruses,[96] and they are essential to the regulation of saltwater and freshwater ecosystems.[97] Most are bacteriophages,[98] which are harmless to plants and animals. They infect and destroy the bacteria in aquatic microbial communities and this is the most important mechanism of recycling carbon in the marine environment. The organic molecules released from the bacterial cells by the viruses stimulate fresh bacterial and algal growth.[99]
Microorganisms constitute more than 90% of the biomass in the sea. It is estimated that viruses kill approximately 20% of this biomass each day and that there are fifteen times as many viruses in the oceans as there are bacteria and archaea. They are mainly responsible for the rapid destruction of harmful algal blooms,[100] which often kill other marine life.[101] The number of viruses in the oceans decreases further offshore and deeper into the water, where there are fewer host organisms.[102]
Their effects are far-reaching; by increasing the amount of respiration in the oceans, viruses are indirectly responsible for reducing the amount of carbon dioxide in the atmosphere by approximately 3 gigatonnes of carbon per year.[102]
Marine mammals are also susceptible to viral infections. In 1988 and 2002, thousands of harbour seals were killed in Europe by phocine distemper virus.[103] Many other viruses, including caliciviruses, herpesviruses, adenoviruses and parvoviruses, circulate in marine mammal populations.[102]
See also
 Medicine portal
Medicine portal Viruses portal
Viruses portal
References
Notes
- King AM, Lefkowitz EJ, Mushegian AR, Adams MJ, Dutilh BE, Gorbalenya AE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert ML, Rubino L, Sabanadzovic S, Sanfaçon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Davison AJ (September 2018). "Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee Taxonomy of Viruses (2018)" (PDF). Archives of Virology. 163 (9): 2601. doi:10.1007/s00705-018-3847-1. PMID 29754305. S2CID 21670772.
- Shors 2017, p. 6
- Collier, Balows & Sussman 1998, p. 3
- Shors 2017, p. 827
- D'Herelle F (2007). "On an invisible microbe antagonistic toward dysenteric bacilli: brief note by Mr. F. D'Herelle, presented by Mr. Roux. 1917". Research in Microbiology. 158 (7): 553–554. doi:10.1016/j.resmic.2007.07.005. PMID 17855060.
- From Nobel Lectures, Physics 1981–1990, (1993) Editor-in-Charge Tore Frängsmyr, Editor Gösta Ekspång, World Scientific Publishing Co., Singapore
- Stanley WM, Loring HS (1936). "The isolation of crystalline tobacco mosaic virus protein from diseased tomato plants". Science. 83 (2143): 85. Bibcode:1936Sci....83...85S. doi:10.1126/science.83.2143.85. PMID 17756690.
- Stanley WM, Lauffer MA (1939). "Disintegration of tobacco mosaic virus in urea solutions". Science. 89 (2311): 345–347. Bibcode:1939Sci....89..345S. doi:10.1126/science.89.2311.345. PMID 17788438.
- Creager AN, Morgan GJ (June 2008). "After the double helix: Rosalind Franklin's research on Tobacco mosaic virus". Isis; an International Review Devoted to the History of Science and Its Cultural Influences. 99 (2): 239–272. doi:10.1086/588626. PMID 18702397. S2CID 25741967.
- Johnson, Ben (25 July 2017). "Rosalind Franklin's contributions to virology". Nature Portfolio Microbiology Community. Retrieved 7 January 2022.
- Goodpasture EW, Woodruff AM, Buddingh GJ (October 1931). "The Cultivation Of Vaccine and other Viruses In The Chorioallantoic Membrane of Chick Embryos". Science. 74 (1919): 371–372. Bibcode:1931Sci....74..371G. doi:10.1126/science.74.1919.371. PMID 17810781.
- Rosen FS (October 2004). "Isolation of poliovirus – John Enders and the Nobel Prize". N. Engl. J. Med. 351 (15): 1481–1483. doi:10.1056/NEJMp048202. PMID 15470207.
- Shors 2017, p. 16
- Collier, Balows & Sussman 1998, pp. 18–19
- Liu Y, Nickle DC, Shriner D, Jensen MA, Learn GH, Mittler JE, Mullins JI (November 2004). "Molecular clock-like evolution of human immunodeficiency virus type 1". Virology. 329 (1): 101–108. doi:10.1016/j.virol.2004.08.014. PMID 15476878.
- Krupovic M, Dooja W, Koonin EV (2019). "Origin of viruses: primordial replicators recruiting capsids from hosts" (PDF). Nature Reviews Microbiology. 17 (7): 449–458. doi:10.1038/s41579-019-0205-6. PMID 31142823. S2CID 169035711.
- Collier, Balows & Sussman 1998, pp. 11–21
- Collier, Balows & Sussman 1998, p. 11
- Collier, Balows & Sussman 1998, pp. 11–12
- Wessner DR (2010). "The Origins of Viruses". Nature Education. 3 (9): 37.
- Nasir A, Kim KM, Caetano-Anollés G (2012). "Viral evolution: Primordial cellular origins and late adaptation to parasitism". Mobile Genetic Elements. 2 (5): 247–252. doi:10.4161/mge.22797. PMC 3575434. PMID 23550145.
- Mahy WJ, Van Regenmortel MH (2009). Desk Encyclopedia of General Virology. Oxford: Academic Press. p. 28. ISBN 978-0-12-375146-1.
- Collier, Balows & Sussman 1998, pp. 33–55
- Rotter ML (August 2001). "Arguments for alcoholic hand disinfection". The Journal of Hospital Infection. 48 (Suppl A): S4–S8. doi:10.1016/s0195-6701(01)90004-0. PMID 11759024.
- Abergel C, Legendre M, Claverie JM (November 2015). "The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus". FEMS Microbiol. Rev. 39 (6): 779–796. doi:10.1093/femsre/fuv037. PMID 26391910.
- Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C (July 2013). "Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes" (PDF). Science. 341 (6143): 281–286. Bibcode:2013Sci...341..281P. doi:10.1126/science.1239181. PMID 23869018. S2CID 16877147.
- Zimmer C (18 July 2013). "Changing View on Viruses: Not So Small After All". The New York Times. Retrieved 20 December 2014.
- Shors 2017, p. 81
- Shors 2017, p. 129
- International Human, Genome Sequencing Consortium (2004). "Finishing the euchromatic sequence of the human genome". Nature. 431 (7011): 931–945. Bibcode:2004Natur.431..931H. doi:10.1038/nature03001. PMID 15496913. S2CID 186242248.
- Shors 2017, pp. 129–31
- Shors 2017, p. 652
- Shors 2017, p. 654
- de Klerk E, 't Hoen PA (March 2015). "Alternative mRNA transcription, processing, and translation: insights from RNA sequencing". Trends in Genetics. 31 (3): 128–139. doi:10.1016/j.tig.2015.01.001. PMID 25648499.
- Collier, Balows & Sussman 1998, pp. 75–82
- Shors 2017, p. 698
- Shors 2017, pp. 6–13
- Shors 2017, pp. 121–123
- Oxford, Kellam & Collier 2016, p. 34–36.
- Oxford, Kellam & Collier 2016, p. 34.
- Okamoto T, Suzuki T, Kusakabe S, Tokunaga M, Hirano J, Miyata Y, Matsuura Y (2017). "Regulation of Apoptosis during Flavivirus Infection". Viruses. 9 (9): 243. doi:10.3390/v9090243. PMC 5618009. PMID 28846635.
- Alwine JC (2008). "Modulation of host cell stress responses by human cytomegalovirus". Human Cytomegalovirus. Curr. Top. Microbiol. Immunol. Current Topics in Microbiology and Immunology. Vol. 325. pp. 263–279. doi:10.1007/978-3-540-77349-8_15. ISBN 978-3-540-77348-1. PMID 18637511.
- Sinclair J (March 2008). "Human cytomegalovirus: Latency and reactivation in the myeloid lineage". J. Clin. Virol. 41 (3): 180–185. doi:10.1016/j.jcv.2007.11.014. PMID 18164651.
- Jordan MC, Jordan GW, Stevens JG, Miller G (June 1984). "Latent herpesviruses of humans". Ann. Intern. Med. 100 (6): 866–880. doi:10.7326/0003-4819-100-6-866. PMID 6326635.
- Sissons JG, Bain M, Wills MR (February 2002). "Latency and reactivation of human cytomegalovirus". J. Infect. 44 (2): 73–77. doi:10.1053/jinf.2001.0948. PMID 12076064. S2CID 24879226.
- Barozzi P, Potenza L, Riva G, Vallerini D, Quadrelli C, Bosco R, Forghieri F, Torelli G, Luppi M (December 2007). "B cells and herpesviruses: a model of lymphoproliferation". Autoimmun Rev. 7 (2): 132–136. doi:10.1016/j.autrev.2007.02.018. PMID 18035323.
- Graham SV (2017). "The human papillomavirus replication cycle, and its links to cancer progression: a comprehensive review". Clinical Science. 131 (17): 2201–2221. doi:10.1042/CS20160786. PMID 28798073.
- Roulston A, Marcellus RC, Branton PE (1999). "Viruses and apoptosis". Annu. Rev. Microbiol. 53: 577–628. doi:10.1146/annurev.micro.53.1.577. PMID 10547702.
- Shors 2017, p. 32
- Shors 2017, p. 271
- Berngruber TW, Froissart R, Choisy M, Gandon S (2013). "Evolution of Virulence in Emerging Epidemics". PLOS Pathogens. 9 (3): e1003209. doi:10.1371/journal.ppat.1003209. PMC 3597519. PMID 23516359.
- Shors 2017, p. 464
- Tanaka J, Akita T, Ko K, Miura Y, Satake M (September 2019). "Countermeasures against viral hepatitis B and C in Japan: An epidemiological point of view". Hepatology Research. 49 (9): 990–1002. doi:10.1111/hepr.13417. PMC 6852166. PMID 31364248.
- Lai CC, Liu YH, Wang CY, Wang YH, Hsueh SC, Yen MY, Ko WC, Hsueh PR (March 2020). "Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths". Journal of Microbiology, Immunology, and Infection = Wei Mian Yu Gan Ran Za Zhi. 53 (3): 404–412. doi:10.1016/j.jmii.2020.02.012. PMC 7128959. PMID 32173241.
- Oxford, Kellam & Collier 2016, p. 63.
- Strand LK (July 2018). "The Terrible Summer of 1952 … When Polio Struck Our Family". Seminars in Pediatric Neurology. 26: 39–44. doi:10.1016/j.spen.2017.04.001. PMID 29961515. S2CID 49640682.
- Moorthy M, Castronovo D, Abraham A, Bhattacharyya S, Gradus S, Gorski J, Naumov YN, Fefferman NH, Naumova EN (October 2012). "Deviations in influenza seasonality: odd coincidence or obscure consequence?". Clinical Microbiology and Infection. 18 (10): 955–962. doi:10.1111/j.1469-0691.2012.03959.x. PMC 3442949. PMID 22958213.
- Barril PA, Fumian TM, Prez VE, Gil PI, Martínez LC, Giordano MO, Masachessi G, Isa MB, Ferreyra LJ, Ré VE, Miagostovich M, Pavan JV, Nates SV (April 2015). "Rotavirus seasonality in urban sewage from Argentina: effect of meteorological variables on the viral load and the genetic diversity". Environmental Research. 138: 409–415. Bibcode:2015ER....138..409B. doi:10.1016/j.envres.2015.03.004. PMID 25777068.
- Durrheim DN, Crowcroft NS, Strebel PM (December 2014). "Measles – The epidemiology of elimination". Vaccine. 32 (51): 6880–6883. doi:10.1016/j.vaccine.2014.10.061. PMID 25444814.
- Mbanzulu KM, Mboera LE, Luzolo FK, Wumba R, Misinzo G, Kimera SI (February 2020). "Mosquito-borne viral diseases in the Democratic Republic of the Congo: a review". Parasites & Vectors. 13 (1): 103. doi:10.1186/s13071-020-3985-7. PMC 7045448. PMID 32103776.
- Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA (March 2020). "Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks". Pathogens (Basel, Switzerland). 9 (3): 186. doi:10.3390/pathogens9030186. PMC 7157630. PMID 32143502.
- Eisinger RW, Fauci AS (March 2018). "Ending the HIV/AIDS Pandemic1". Emerging Infectious Diseases. 24 (3): 413–416. doi:10.3201/eid2403.171797. PMC 5823353. PMID 29460740.
- Qin Y, Zhao MJ, Tan YY, Li XQ, Zheng JD, Peng ZB, Feng LZ (August 2018). "[History of influenza pandemics in China during the past century]". Zhonghua Liu Xing Bing Xue Za Zhi = Zhonghua Liuxingbingxue Zazhi (in Chinese). 39 (8): 1028–1031. doi:10.3760/cma.j.issn.0254-6450.2018.08.003. PMID 30180422.
- Nishiyama Y, Matsukuma S, Matsumura T, Kanatani Y, Saito T (April 2015). "Preparedness for a smallpox pandemic in Japan: public health perspectives". Disaster Medicine and Public Health Preparedness. 9 (2): 220–223. doi:10.1017/dmp.2014.157. PMID 26060873. S2CID 37149836.
- Houghton F (2019). "Geography, global pandemics & air travel: Faster, fuller, further & more frequent". Journal of Infection and Public Health. 12 (3): 448–449. doi:10.1016/j.jiph.2019.02.020. PMC 7129534. PMID 30878442.
- "Virology Journal". Virology Journal.
- Weiss SR, Leibowitz JL (2011). Coronavirus pathogenesis. Advances in Virus Research. Vol. 81. pp. 85–164. doi:10.1016/B978-0-12-385885-6.00009-2. ISBN 978-0-12-385885-6. PMC 7149603. PMID 22094080.
- Wong AT, Chen H, Liu SH, Hsu EK, Luk KS, Lai CK, Chan RF, Tsang OT, Choi KW, Kwan YW, Tong AY, Cheng VC, Tsang DC (May 2017). "From SARS to Avian Influenza Preparedness in Hong Kong". Clinical Infectious Diseases. 64 (suppl_2): S98–S104. doi:10.1093/cid/cix123. PMID 28475794.
- Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) (PDF) (Report). World Health Organization (WHO). 16–24 February 2020. Retrieved 21 March 2020.
- Deng SQ, Peng HJ (February 2020). "Characteristics of and Public Health Responses to the Coronavirus Disease 2019 Outbreak in China". Journal of Clinical Medicine. 9 (2): 575. doi:10.3390/jcm9020575. PMC 7074453. PMID 32093211.
- Han Q, Lin Q, Jin S, You L (February 2020). "Coronavirus 2019-nCoV: A brief perspective from the front line". The Journal of Infection. 80 (4): 373–377. doi:10.1016/j.jinf.2020.02.010. PMC 7102581. PMID 32109444.
- Londoño E, Ortiz A (16 March 2020). "Coronavirus Travel Restrictions, Across the Globe" – via NYTimes.com.
- "US takes more big pandemic response steps; Europe COVID-19 cases soar". CIDRAP.
- Shors 2017, p. 822
- Shors 2017, pp. 802–803
- Oxford, Kellam & Collier 2016, p. 19.
- Shors 2017, p. 803
- Shors 2017, pp. 116–117
- Shors 2017, pp. 225–233
- Garcia-Ruiz H (2018). "Susceptibility Genes to Plant Viruses". Viruses. 10 (9): 484. doi:10.3390/v10090484. PMC 6164914. PMID 30201857.
- Shors 2017, p. 812
- Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP (2005). "Mechanisms of plant resistance to viruses". Nature Reviews Microbiology. 3 (10): 789–798. doi:10.1038/nrmicro1239. PMID 16132037. S2CID 27311732.
- Horvath P, Barrangou R (January 2010). "CRISPR/Cas, the immune system of bacteria and archaea" (PDF). Science. 327 (5962): 167–170. Bibcode:2010Sci...327..167H. doi:10.1126/science.1179555. PMID 20056882. S2CID 17960960. Archived from the original (PDF) on 27 March 2020.
- Shors 2017, pp. 237–255
- Small JC, Ertl HC (2011). "Viruses – from pathogens to vaccine carriers". Current Opinion in Virology. 1 (4): 241–245. doi:10.1016/j.coviro.2011.07.009. PMC 3190199. PMID 22003377.
- Burakova Y, Madera R, McVey S, Schlup JR, Shi J (2018). "Adjuvants for Animal Vaccines". Viral Immunology. 31 (1): 11–22. doi:10.1089/vim.2017.0049. PMID 28618246.
- Shors 2017, p. 237
- Thomssen R (1975). "Live attenuated versus killed virus vaccines". Monographs in Allergy. 9: 155–176. PMID 1090805.
- Shors 2017, p. 238
- Shors 2017, pp. 514–515
- Shors 2017, p. 514
- Shors 2017, p. 568
- Falade-Nwulia O, Suarez-Cuervo C, Nelson DR, Fried MW, Segal JB, Sulkowski MS (May 2017). "Oral Direct-Acting Agent Therapy for Hepatitis C Virus Infection: A Systematic Review". Annals of Internal Medicine. 166 (9): 637–648. doi:10.7326/M16-2575. PMC 5486987. PMID 28319996.
- Paul N, Han SH (June 2011). "Combination Therapy for Chronic Hepatitis B: Current Indications". Curr Hepat Rep. 10 (2): 98–105. doi:10.1007/s11901-011-0095-1. PMC 3085106. PMID 21654909.
- Koonin EV, Senkevich TG, Dolja VV (September 2006). "The ancient Virus World and evolution of cells". Biol. Direct. 1: 29. doi:10.1186/1745-6150-1-29. PMC 1594570. PMID 16984643.
- Dávila-Ramos S, Castelán-Sánchez HG, Martínez-Ávila L, Sánchez-Carbente MD, Peralta R, Hernández-Mendoza A, Dobson AD, Gonzalez RA, Pastor N, Batista-García RA (2019). "A Review on Viral Metagenomics in Extreme Environments". Frontiers in Microbiology. 10: 2403. doi:10.3389/fmicb.2019.02403. PMC 6842933. PMID 31749771.
- Shors 2017, p. 5
- Breitbart M, Bonnain C, Malki K, Sawaya NA (July 2018). "Phage puppet masters of the marine microbial realm". Nature Microbiology. 3 (7): 754–766. doi:10.1038/s41564-018-0166-y. PMID 29867096. S2CID 46927784.
- Shors 2017, pp. 25–26
- Suttle CA (September 2005). "Viruses in the sea". Nature. 437 (7057): 356–361. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. PMID 16163346. S2CID 4370363.
- "Harmful Algal Blooms: Red Tide: Home | CDC HSB". www.cdc.gov. Retrieved 23 August 2009.
- Suttle CA (October 2007). "Marine viruses – major players in the global ecosystem". Nat. Rev. Microbiol. 5 (10): 801–812. doi:10.1038/nrmicro1750. PMID 17853907. S2CID 4658457.
- Hall A, Jepson P, Goodman S, Harkonen T (2006). "Phocine distemper virus in the North and European Seas – Data and models, nature and nurture". Biological Conservation. 131 (2): 221–229. doi:10.1016/j.biocon.2006.04.008.
Bibliography
- Collier L, Balows A, Sussman M, eds. (1998). Topley & Wilson's Microbiology and Microbial Infections. Vol. 1, Virology (9th ed.). Arnold. ISBN 0-340-66316-2.
- Oxford, John; Kellam, Paul; Collier, Leslie (2016). Human Virology. Oxford: Oxford University Press. ISBN 978-0-19-871468-2. OCLC 968152575.
- Shors T (2017). Understanding Viruses. Jones and Bartlett Publishers. ISBN 978-1284025927.
External links
| Library resources about Virus |
- Virus Pathogen Resource – Genomic and other research data about human pathogenic viruses
- Influenza Research Database – Genomic and other research data about influenza viruses