Bacteria
Bacteria (/bækˈtɪəriə/ (![]() listen); singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
listen); singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
| Bacteria | |
|---|---|
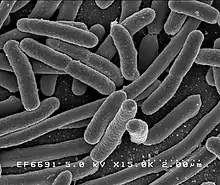 | |
| Scanning electron micrograph of Escherichia coli rods | |
| Scientific classification | |
| Domain: | Bacteria Woese et al. 1990 |
| Phyla | |
| Synonyms | |
| |
Humans and most other animals carry millions of bacteria. Most are in the gut, and there are many on the skin. Most of the bacteria in and on the body are harmless or rendered so by the protective effects of the immune system, and many are beneficial,[2] particularly the ones in the gut. However, several species of bacteria are pathogenic and cause infectious diseases, including cholera, syphilis, anthrax, leprosy, tuberculosis, tetanus and bubonic plague. The most common fatal bacterial diseases are respiratory infections. Antibiotics are used to treat bacterial infections and are also used in farming, making antibiotic resistance a growing problem. Bacteria are important in sewage treatment and the breakdown of oil spills, the production of cheese and yogurt through fermentation, the recovery of gold, palladium, copper and other metals in the mining sector, as well as in biotechnology, and the manufacture of antibiotics and other chemicals.
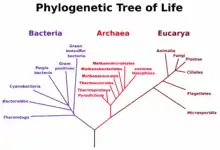
Once regarded as plants constituting the class Schizomycetes ("fission fungi"), bacteria are now classified as prokaryotes. Unlike cells of animals and other eukaryotes, bacterial cells do not contain a nucleus and rarely harbour membrane-bound organelles. Although the term bacteria traditionally included all prokaryotes, the scientific classification changed after the discovery in the 1990s that prokaryotes consist of two very different groups of organisms that evolved from an ancient common ancestor. These evolutionary domains are called Bacteria and Archaea.[3]
Etymology
The word bacteria is the plural of the New Latin bacterium, which is the latinisation of the Ancient Greek βακτήριον (baktḗrion),[4] the diminutive of βακτηρία (baktēría), meaning "staff, cane",[5] because the first ones to be discovered were rod-shaped.[6][7]
Origin and early evolution
The ancestors of bacteria were unicellular microorganisms that were the first forms of life to appear on Earth, about 4 billion years ago.[9] For about 3 billion years, most organisms were microscopic, and bacteria and archaea were the dominant forms of life.[10][11][12] Although bacterial fossils exist, such as stromatolites, their lack of distinctive morphology prevents them from being used to examine the history of bacterial evolution, or to date the time of origin of a particular bacterial species. However, gene sequences can be used to reconstruct the bacterial phylogeny, and these studies indicate that bacteria diverged first from the archaeal/eukaryotic lineage.[13] The most recent common ancestor of bacteria and archaea was probably a hyperthermophile that lived about 2.5 billion–3.2 billion years ago.[14][15][16] The earliest life on land may have been bacteria some 3.22 billion years ago.[17]
Bacteria were also involved in the second great evolutionary divergence, that of the archaea and eukaryotes.[18][19] Here, eukaryotes resulted from the entering of ancient bacteria into endosymbiotic associations with the ancestors of eukaryotic cells, which were themselves possibly related to the Archaea.[20][21] This involved the engulfment by proto-eukaryotic cells of alphaproteobacterial symbionts to form either mitochondria or hydrogenosomes, which are still found in all known Eukarya (sometimes in highly reduced form, e.g. in ancient "amitochondrial" protozoa). Later, some eukaryotes that already contained mitochondria also engulfed cyanobacteria-like organisms, leading to the formation of chloroplasts in algae and plants. This is known as primary endosymbiosis.[22]
Habitat
Bacteria are ubiquitous, living in every possible habitat on the planet including soil, underwater, deep in Earth's crust and even such extreme environments as acidic hot springs and radioactive waste.[23][24] There are approximately 2×1030 bacteria on Earth,[25] forming a biomass that is only exceeded by plants.[26] They are abundant in lakes and oceans, in arctic ice, and geothermal springs[27] where they provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy.[28] They live on and in plants and animals. Most do not cause diseases, are beneficial to their environments, and are essential for life.[2][29] The soil is a rich source of bacteria and a few grams contain around a thousand million of them. They are all essential to soil ecology, breaking down toxic waste and recycling nutrients. They are even found in the atmosphere and one cubic metre of air holds around one hundred million bacterial cells. The oceans and seas harbour around 3 x 1026 bacteria which provide up to 50% of the oxygen humans breathe.[30] Only around 2% of bacterial species have been fully studied.[31]
| Habitat | Species | Reference |
|---|---|---|
| Cold (minus 15 °C Antarctica) | Cryptoendoliths | [32] |
| Hot (70–100 °C geysers) | Thermus aquaticus | [31] |
| Radiation, 5MRad | Deinococcus radiodurans | [32] |
| Saline, 47% salt (Dead Sea, Great Salt Lake) | several species | ,[31][32] |
| Acid pH 3 | several species | [23] |
| Alkaline pH 12.8 | betaproteobacteria | [32] |
| Space (6 years on a NASA satellite) | Bacillus subtilis | [32] |
| 3.2 km underground | several species | [32] |
| High pressure (Mariana Trench – 1200 atm) | Moritella, Shewanella and others | [32] |
Morphology
Size. Bacteria display a wide diversity of shapes and sizes. Bacterial cells are about one-tenth the size of eukaryotic cells and are typically 0.5–5.0 micrometres in length. However, a few species are visible to the unaided eye—for example, Thiomargarita namibiensis is up to half a millimetre long,[33] Epulopiscium fishelsoni reaches 0.7 mm,[34] and Thiomargarita magnifica can reach even 2 cm in length, which is 50 times larger than other known bacteria.[35][36] Among the smallest bacteria are members of the genus Mycoplasma, which measure only 0.3 micrometres, as small as the largest viruses.[37] Some bacteria may be even smaller, but these ultramicrobacteria are not well-studied.[38]
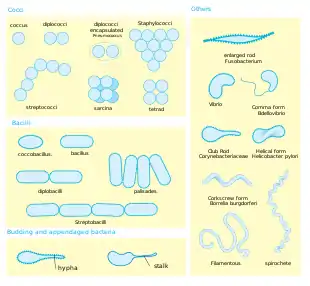
Shape. Most bacterial species are either spherical, called cocci (singular coccus, from Greek kókkos, grain, seed), or rod shaped, called bacilli (sing. bacillus, from Latin baculus, stick).[39] Some bacteria, called vibrio, are shaped like slightly curved rods or comma shaped; others can be spiral shaped, called spirilla, or tightly coiled, called spirochaetes. A small number of other unusual shapes have been described, such as star-shaped bacteria.[40] This wide variety of shapes is determined by the bacterial cell wall and cytoskeleton, and is important because it can influence the ability of bacteria to acquire nutrients, attach to surfaces, swim through liquids and escape predators.[41][42]

Multicellularity. Most bacterial species exist simply as single cells; others associate in characteristic patterns: Neisseria forms diploids (pairs), streptococci form chains, and staphylococci group together in "bunch of grapes" clusters. Bacteria can also group to form larger multicellular structures, such as the elongated filaments of Actinomycetota species, the aggregates of Myxobacteria species, and the complex hyphae of Streptomyces species.[44] These multicellular structures are often only seen in certain conditions. For example, when starved of amino acids, myxobacteria detect surrounding cells in a process known as quorum sensing, migrate towards each other, and aggregate to form fruiting bodies up to 500 micrometres long and containing approximately 100,000 bacterial cells.[45] In these fruiting bodies, the bacteria perform separate tasks; for example, about one in ten cells migrate to the top of a fruiting body and differentiate into a specialised dormant state called a myxospore, which is more resistant to drying and other adverse environmental conditions.[46]
Biofilms. Bacteria often attach to surfaces and form dense aggregations called biofilms,[47] and larger formations known as microbial mats.[48] These biofilms and mats can range from a few micrometres in thickness to up to half a metre in depth, and may contain multiple species of bacteria, protists and archaea. Bacteria living in biofilms display a complex arrangement of cells and extracellular components, forming secondary structures, such as microcolonies, through which there are networks of channels to enable better diffusion of nutrients.[49][50] In natural environments, such as soil or the surfaces of plants, the majority of bacteria are bound to surfaces in biofilms.[51] Biofilms are also important in medicine, as these structures are often present during chronic bacterial infections or in infections of implanted medical devices, and bacteria protected within biofilms are much harder to kill than individual isolated bacteria.[52]
Cellular structure
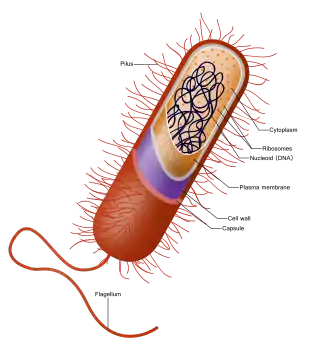
Intracellular structures
The bacterial cell is surrounded by a cell membrane, which is made primarily of phospholipids. This membrane encloses the contents of the cell and acts as a barrier to hold nutrients, proteins and other essential components of the cytoplasm within the cell.[53] Unlike eukaryotic cells, bacteria usually lack large membrane-bound structures in their cytoplasm such as a nucleus, mitochondria, chloroplasts and the other organelles present in eukaryotic cells.[54] However, some bacteria have protein-bound organelles in the cytoplasm which compartmentalize aspects of bacterial metabolism,[55][56] such as the carboxysome.[57] Additionally, bacteria have a multi-component cytoskeleton to control the localisation of proteins and nucleic acids within the cell, and to manage the process of cell division.[58][59][60]
Many important biochemical reactions, such as energy generation, occur due to concentration gradients across membranes, creating a potential difference analogous to a battery. The general lack of internal membranes in bacteria means these reactions, such as electron transport, occur across the cell membrane between the cytoplasm and the outside of the cell or periplasm.[61] However, in many photosynthetic bacteria the plasma membrane is highly folded and fills most of the cell with layers of light-gathering membrane.[62] These light-gathering complexes may even form lipid-enclosed structures called chlorosomes in green sulfur bacteria.[63]
Bacteria do not have a membrane-bound nucleus, and their genetic material is typically a single circular bacterial chromosome of DNA located in the cytoplasm in an irregularly shaped body called the nucleoid.[64] The nucleoid contains the chromosome with its associated proteins and RNA. Like all other organisms, bacteria contain ribosomes for the production of proteins, but the structure of the bacterial ribosome is different from that of eukaryotes and archaea.[65]
Some bacteria produce intracellular nutrient storage granules, such as glycogen,[66] polyphosphate,[67] sulfur[68] or polyhydroxyalkanoates.[69] Bacteria such as the photosynthetic cyanobacteria, produce internal gas vacuoles, which they use to regulate their buoyancy, allowing them to move up or down into water layers with different light intensities and nutrient levels.[70]
Extracellular structures
Around the outside of the cell membrane is the cell wall. Bacterial cell walls are made of peptidoglycan (also called murein), which is made from polysaccharide chains cross-linked by peptides containing D-amino acids.[71] Bacterial cell walls are different from the cell walls of plants and fungi, which are made of cellulose and chitin, respectively.[72] The cell wall of bacteria is also distinct from that of achaea, which do not contain peptidoglycan. The cell wall is essential to the survival of many bacteria, and the antibiotic penicillin (produced by a fungus called Penicillium) is able to kill bacteria by inhibiting a step in the synthesis of peptidoglycan.[72]
There are broadly speaking two different types of cell wall in bacteria, that classify bacteria into Gram-positive bacteria and Gram-negative bacteria. The names originate from the reaction of cells to the Gram stain, a long-standing test for the classification of bacterial species.[73]
Gram-positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids. In contrast, Gram-negative bacteria have a relatively thin cell wall consisting of a few layers of peptidoglycan surrounded by a second lipid membrane containing lipopolysaccharides and lipoproteins. Most bacteria have the Gram-negative cell wall, and only members of the Bacillota group and actinomycetota (previously known as the low G+C and high G+C Gram-positive bacteria, respectively) have the alternative Gram-positive arrangement.[74] These differences in structure can produce differences in antibiotic susceptibility; for instance, vancomycin can kill only Gram-positive bacteria and is ineffective against Gram-negative pathogens, such as Haemophilus influenzae or Pseudomonas aeruginosa.[75] Some bacteria have cell wall structures that are neither classically Gram-positive or Gram-negative. This includes clinically important bacteria such as mycobacteria which have a thick peptidoglycan cell wall like a Gram-positive bacterium, but also a second outer layer of lipids.[76]
In many bacteria, an S-layer of rigidly arrayed protein molecules covers the outside of the cell.[77] This layer provides chemical and physical protection for the cell surface and can act as a macromolecular diffusion barrier. S-layers have diverse functions and are known to act as virulence factors in Campylobacter species and contain surface enzymes in Bacillus stearothermophilus.[78][79]

Flagella are rigid protein structures, about 20 nanometres in diameter and up to 20 micrometres in length, that are used for motility. Flagella are driven by the energy released by the transfer of ions down an electrochemical gradient across the cell membrane.[80]
Fimbriae (sometimes called "attachment pili") are fine filaments of protein, usually 2–10 nanometres in diameter and up to several micrometres in length. They are distributed over the surface of the cell, and resemble fine hairs when seen under the electron microscope.[81] Fimbriae are believed to be involved in attachment to solid surfaces or to other cells, and are essential for the virulence of some bacterial pathogens.[82] Pili (sing. pilus) are cellular appendages, slightly larger than fimbriae, that can transfer genetic material between bacterial cells in a process called conjugation where they are called conjugation pili or sex pili (see bacterial genetics, below).[83] They can also generate movement where they are called type IV pili.[84]
Glycocalyx is produced by many bacteria to surround their cells,[85] and varies in structural complexity: ranging from a disorganised slime layer of extracellular polymeric substances to a highly structured capsule. These structures can protect cells from engulfment by eukaryotic cells such as macrophages (part of the human immune system).[86] They can also act as antigens and be involved in cell recognition, as well as aiding attachment to surfaces and the formation of biofilms.[87]
The assembly of these extracellular structures is dependent on bacterial secretion systems. These transfer proteins from the cytoplasm into the periplasm or into the environment around the cell. Many types of secretion systems are known and these structures are often essential for the virulence of pathogens, so are intensively studied.[87]
Endospores
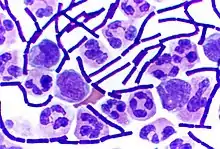
Some genera of Gram-positive bacteria, such as Bacillus, Clostridium, Sporohalobacter, Anaerobacter, and Heliobacterium, can form highly resistant, dormant structures called endospores.[89] Endospores develop within the cytoplasm of the cell; generally a single endospore develops in each cell.[90] Each endospore contains a core of DNA and ribosomes surrounded by a cortex layer and protected by a multilayer rigid coat composed of peptidoglycan and a variety of proteins.[90]
Endospores show no detectable metabolism and can survive extreme physical and chemical stresses, such as high levels of UV light, gamma radiation, detergents, disinfectants, heat, freezing, pressure, and desiccation.[91] In this dormant state, these organisms may remain viable for millions of years,[92][93][94] and endospores even allow bacteria to survive exposure to the vacuum and radiation in space, possibly bacteria could be distributed throughout the Universe by space dust, meteoroids, asteroids, comets, planetoids or via directed panspermia.[95][96] Endospore-forming bacteria can also cause disease: for example, anthrax can be contracted by the inhalation of Bacillus anthracis endospores, and contamination of deep puncture wounds with Clostridium tetani endospores causes tetanus, which like botulism is caused by a toxin released by the bacteria that grow from the spores.[97] Clostridioides difficile infection, which is a problem in healthcare settings is also caused by spore-forming bacteria.[98]
Metabolism
Bacteria exhibit an extremely wide variety of metabolic types.[99] The distribution of metabolic traits within a group of bacteria has traditionally been used to define their taxonomy, but these traits often do not correspond with modern genetic classifications.[100] Bacterial metabolism is classified into nutritional groups on the basis of three major criteria: the source of energy, the electron donors used, and the source of carbon used for growth.[101]
Bacteria either derive energy from light using photosynthesis (called phototrophy), or by breaking down chemical compounds using oxidation (called chemotrophy).[102] Chemotrophs use chemical compounds as a source of energy by transferring electrons from a given electron donor to a terminal electron acceptor in a redox reaction. This reaction releases energy that can be used to drive metabolism. Chemotrophs are further divided by the types of compounds they use to transfer electrons. Bacteria that use inorganic compounds such as hydrogen, carbon monoxide, or ammonia as sources of electrons are called lithotrophs, while those that use organic compounds are called organotrophs.[102] The compounds used to receive electrons are also used to classify bacteria: aerobic organisms use oxygen as the terminal electron acceptor, while anaerobic organisms use other compounds such as nitrate, sulfate, or carbon dioxide.[102]
Many bacteria get their carbon from other organic carbon, called heterotrophy. Others such as cyanobacteria and some purple bacteria are autotrophic, meaning that they obtain cellular carbon by fixing carbon dioxide.[103] In unusual circumstances, the gas methane can be used by methanotrophic bacteria as both a source of electrons and a substrate for carbon anabolism.[104]
| Nutritional type | Source of energy | Source of carbon | Examples |
|---|---|---|---|
| Phototrophs | Sunlight | Organic compounds (photoheterotrophs) or carbon fixation (photoautotrophs) | Cyanobacteria, Green sulfur bacteria, Chloroflexota, or Purple bacteria |
| Lithotrophs | Inorganic compounds | Organic compounds (lithoheterotrophs) or carbon fixation (lithoautotrophs) | Thermodesulfobacteriota, Hydrogenophilaceae, or Nitrospirota |
| Organotrophs | Organic compounds | Organic compounds (chemoheterotrophs) or carbon fixation (chemoautotrophs) | Bacillus, Clostridium, or Enterobacteriaceae |
In many ways, bacterial metabolism provides traits that are useful for ecological stability and for human society. One example is that some bacteria called diazotrophs have the ability to fix nitrogen gas using the enzyme nitrogenase.[105] This environmentally important trait can be found in bacteria of most metabolic types listed above.[106] This leads to the ecologically important processes of denitrification, sulfate reduction, and acetogenesis, respectively.[107] Bacterial metabolic processes are also important in biological responses to pollution; for example, sulfate-reducing bacteria are largely responsible for the production of the highly toxic forms of mercury (methyl- and dimethylmercury) in the environment.[108] Non-respiratory anaerobes use fermentation to generate energy and reducing power, secreting metabolic by-products (such as ethanol in brewing) as waste. Facultative anaerobes can switch between fermentation and different terminal electron acceptors depending on the environmental conditions in which they find themselves.[109]
Growth and reproduction
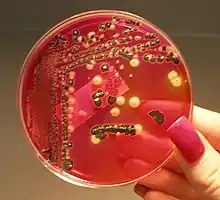

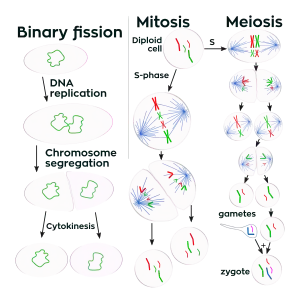
Unlike in multicellular organisms, increases in cell size (cell growth) and reproduction by cell division are tightly linked in unicellular organisms. Bacteria grow to a fixed size and then reproduce through binary fission, a form of asexual reproduction.[111] Under optimal conditions, bacteria can grow and divide extremely rapidly, and some bacterial populations can double as quickly as every 17 minutes.[112] In cell division, two identical clone daughter cells are produced. Some bacteria, while still reproducing asexually, form more complex reproductive structures that help disperse the newly formed daughter cells. Examples include fruiting body formation by myxobacteria and aerial hyphae formation by Streptomyces species, or budding. Budding involves a cell forming a protrusion that breaks away and produces a daughter cell.[113]
In the laboratory, bacteria are usually grown using solid or liquid media.[114] Solid growth media, such as agar plates, are used to isolate pure cultures of a bacterial strain. However, liquid growth media are used when the measurement of growth or large volumes of cells are required. Growth in stirred liquid media occurs as an even cell suspension, making the cultures easy to divide and transfer, although isolating single bacteria from liquid media is difficult. The use of selective media (media with specific nutrients added or deficient, or with antibiotics added) can help identify specific organisms.[115]
Most laboratory techniques for growing bacteria use high levels of nutrients to produce large amounts of cells cheaply and quickly.[114] However, in natural environments, nutrients are limited, meaning that bacteria cannot continue to reproduce indefinitely. This nutrient limitation has led the evolution of different growth strategies (see r/K selection theory). Some organisms can grow extremely rapidly when nutrients become available, such as the formation of algal and cyanobacterial blooms that often occur in lakes during the summer.[116] Other organisms have adaptations to harsh environments, such as the production of multiple antibiotics by streptomyces that inhibit the growth of competing microorganisms.[117] In nature, many organisms live in communities (e.g., biofilms) that may allow for increased supply of nutrients and protection from environmental stresses.[51] These relationships can be essential for growth of a particular organism or group of organisms (syntrophy).[118]
Bacterial growth follows four phases. When a population of bacteria first enter a high-nutrient environment that allows growth, the cells need to adapt to their new environment. The first phase of growth is the lag phase, a period of slow growth when the cells are adapting to the high-nutrient environment and preparing for fast growth. The lag phase has high biosynthesis rates, as proteins necessary for rapid growth are produced.[119][120] The second phase of growth is the logarithmic phase, also known as the exponential phase. The log phase is marked by rapid exponential growth. The rate at which cells grow during this phase is known as the growth rate (k), and the time it takes the cells to double is known as the generation time (g). During log phase, nutrients are metabolised at maximum speed until one of the nutrients is depleted and starts limiting growth. The third phase of growth is the stationary phase and is caused by depleted nutrients. The cells reduce their metabolic activity and consume non-essential cellular proteins. The stationary phase is a transition from rapid growth to a stress response state and there is increased expression of genes involved in DNA repair, antioxidant metabolism and nutrient transport.[121] The final phase is the death phase where the bacteria run out of nutrients and die.[122]
Genetics
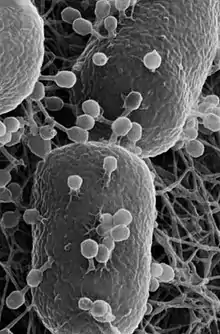
Most bacteria have a single circular chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Carsonella ruddii,[124] to 12,200,000 base pairs (12.2 Mbp) in the soil-dwelling bacteria Sorangium cellulosum.[125] There are many exceptions to this, for example some Streptomyces and Borrelia species contain a single linear chromosome,[126][127] while some Vibrio species contain more than one chromosome.[128] Bacteria can also contain plasmids, small extra-chromosomal molecules of DNA that may contain genes for various useful functions such as antibiotic resistance, metabolic capabilities, or various virulence factors.[129]
Bacteria genomes usually encode a few hundred to a few thousand genes. The genes in bacterial genomes are usually a single continuous stretch of DNA and although several different types of introns do exist in bacteria, these are much rarer than in eukaryotes.[130]
Bacteria, as asexual organisms, inherit an identical copy of the parent's genomes and are clonal. However, all bacteria can evolve by selection on changes to their genetic material DNA caused by genetic recombination or mutations. Mutations come from errors made during the replication of DNA or from exposure to mutagens. Mutation rates vary widely among different species of bacteria and even among different clones of a single species of bacteria.[131] Genetic changes in bacterial genomes come from either random mutation during replication or "stress-directed mutation", where genes involved in a particular growth-limiting process have an increased mutation rate.[132]
Some bacteria also transfer genetic material between cells. This can occur in three main ways. First, bacteria can take up exogenous DNA from their environment, in a process called transformation.[133] Many bacteria can naturally take up DNA from the environment, while others must be chemically altered in order to induce them to take up DNA.[134] The development of competence in nature is usually associated with stressful environmental conditions, and seems to be an adaptation for facilitating repair of DNA damage in recipient cells.[135] The second way bacteria transfer genetic material is by transduction, when the integration of a bacteriophage introduces foreign DNA into the chromosome. Many types of bacteriophage exist, some simply infect and lyse their host bacteria, while others insert into the bacterial chromosome.[136] Bacteria resist phage infection through restriction modification systems that degrade foreign DNA,[137] and a system that uses CRISPR sequences to retain fragments of the genomes of phage that the bacteria have come into contact with in the past, which allows them to block virus replication through a form of RNA interference.[138][139] The third method of gene transfer is conjugation, whereby DNA is transferred through direct cell contact. In ordinary circumstances, transduction, conjugation, and transformation involve transfer of DNA between individual bacteria of the same species, but occasionally transfer may occur between individuals of different bacterial species and this may have significant consequences, such as the transfer of antibiotic resistance.[140][141] In such cases, gene acquisition from other bacteria or the environment is called horizontal gene transfer and may be common under natural conditions.[142]
Behaviour
Movement
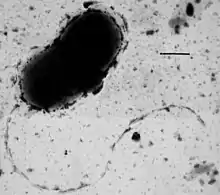
Many bacteria are motile (able to move themselves) and do so using a variety of mechanisms. The best studied of these are flagella, long filaments that are turned by a motor at the base to generate propeller-like movement.[143] The bacterial flagellum is made of about 20 proteins, with approximately another 30 proteins required for its regulation and assembly.[143] The flagellum is a rotating structure driven by a reversible motor at the base that uses the electrochemical gradient across the membrane for power.[144]
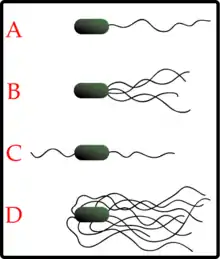
Bacteria can use flagella in different ways to generate different kinds of movement. Many bacteria (such as E. coli) have two distinct modes of movement: forward movement (swimming) and tumbling. The tumbling allows them to reorient and makes their movement a three-dimensional random walk.[145] Bacterial species differ in the number and arrangement of flagella on their surface; some have a single flagellum (monotrichous), a flagellum at each end (amphitrichous), clusters of flagella at the poles of the cell (lophotrichous), while others have flagella distributed over the entire surface of the cell (peritrichous). The flagella of a unique group of bacteria, the spirochaetes, are found between two membranes in the periplasmic space. They have a distinctive helical body that twists about as it moves.[143]
Two other types of bacterial motion are called twitching motility that relies on a structure called the type IV pilus,[146] and gliding motility, that uses other mechanisms. In twitching motility, the rod-like pilus extends out from the cell, binds some substrate, and then retracts, pulling the cell forward.[147]
Motile bacteria are attracted or repelled by certain stimuli in behaviours called taxes: these include chemotaxis, phototaxis, energy taxis, and magnetotaxis.[148][149][150] In one peculiar group, the myxobacteria, individual bacteria move together to form waves of cells that then differentiate to form fruiting bodies containing spores.[46] The myxobacteria move only when on solid surfaces, unlike E. coli, which is motile in liquid or solid media.[151]
Several Listeria and Shigella species move inside host cells by usurping the cytoskeleton, which is normally used to move organelles inside the cell. By promoting actin polymerisation at one pole of their cells, they can form a kind of tail that pushes them through the host cell's cytoplasm.[152]
Communication
A few bacteria have chemical systems that generate light. This bioluminescence often occurs in bacteria that live in association with fish, and the light probably serves to attract fish or other large animals.[153]
Bacteria often function as multicellular aggregates known as biofilms, exchanging a variety of molecular signals for inter-cell communication, and engaging in coordinated multicellular behaviour.[154][155]
The communal benefits of multicellular cooperation include a cellular division of labour, accessing resources that cannot effectively be used by single cells, collectively defending against antagonists, and optimising population survival by differentiating into distinct cell types.[154] For example, bacteria in biofilms can have more than 500 times increased resistance to antibacterial agents than individual "planktonic" bacteria of the same species.[155]
One type of inter-cellular communication by a molecular signal is called quorum sensing, which serves the purpose of determining whether there is a local population density that is sufficiently high that it is productive to invest in processes that are only successful if large numbers of similar organisms behave similarly, as in excreting digestive enzymes or emitting light.[156][157]
Quorum sensing allows bacteria to coordinate gene expression, and enables them to produce, release and detect autoinducers or pheromones which accumulate with the growth in cell population.[158]
Classification and identification
Classification seeks to describe the diversity of bacterial species by naming and grouping organisms based on similarities. Bacteria can be classified on the basis of cell structure, cellular metabolism or on differences in cell components, such as DNA, fatty acids, pigments, antigens and quinones.[115] While these schemes allowed the identification and classification of bacterial strains, it was unclear whether these differences represented variation between distinct species or between strains of the same species. This uncertainty was due to the lack of distinctive structures in most bacteria, as well as lateral gene transfer between unrelated species.[160] Due to lateral gene transfer, some closely related bacteria can have very different morphologies and metabolisms. To overcome this uncertainty, modern bacterial classification emphasises molecular systematics, using genetic techniques such as guanine cytosine ratio determination, genome-genome hybridisation, as well as sequencing genes that have not undergone extensive lateral gene transfer, such as the rRNA gene.[161] Classification of bacteria is determined by publication in the International Journal of Systematic Bacteriology,[162] and Bergey's Manual of Systematic Bacteriology.[163] The International Committee on Systematic Bacteriology (ICSB) maintains international rules for the naming of bacteria and taxonomic categories and for the ranking of them in the International Code of Nomenclature of Bacteria.[164]
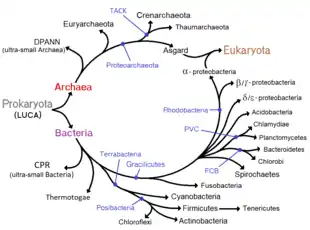
Historically, bacteria were considered a part of the Plantae, the Plant kingdom, and were called "Schizomycetes" (fission-fungi).[165] For this reason, collective bacteria and other microorganisms in a host are often called "flora".[166] The term "bacteria" was traditionally applied to all microscopic, single-cell prokaryotes. However, molecular systematics showed prokaryotic life to consist of two separate domains, originally called Eubacteria and Archaebacteria, but now called Bacteria and Archaea that evolved independently from an ancient common ancestor.[3] The archaea and eukaryotes are more closely related to each other than either is to the bacteria. These two domains, along with Eukarya, are the basis of the three-domain system, which is currently the most widely used classification system in microbiology.[167] However, due to the relatively recent introduction of molecular systematics and a rapid increase in the number of genome sequences that are available, bacterial classification remains a changing and expanding field.[168][169] For example, Cavalier-Smith argued that the Archaea and Eukaryotes evolved from Gram-positive bacteria.[170]
The identification of bacteria in the laboratory is particularly relevant in medicine, where the correct treatment is determined by the bacterial species causing an infection. Consequently, the need to identify human pathogens was a major impetus for the development of techniques to identify bacteria.[171]
The Gram stain, developed in 1884 by Hans Christian Gram, characterises bacteria based on the structural characteristics of their cell walls.[172][73] The thick layers of peptidoglycan in the "Gram-positive" cell wall stain purple, while the thin "Gram-negative" cell wall appears pink.[172] By combining morphology and Gram-staining, most bacteria can be classified as belonging to one of four groups (Gram-positive cocci, Gram-positive bacilli, Gram-negative cocci and Gram-negative bacilli). Some organisms are best identified by stains other than the Gram stain, particularly mycobacteria or Nocardia, which show acid fastness on Ziehl–Neelsen or similar stains.[173] Other organisms may need to be identified by their growth in special media, or by other techniques, such as serology.[174]
Culture techniques are designed to promote the growth and identify particular bacteria, while restricting the growth of the other bacteria in the sample.[175] Often these techniques are designed for specific specimens; for example, a sputum sample will be treated to identify organisms that cause pneumonia, while stool specimens are cultured on selective media to identify organisms that cause diarrhea, while preventing growth of non-pathogenic bacteria. Specimens that are normally sterile, such as blood, urine or spinal fluid, are cultured under conditions designed to grow all possible organisms.[115][176] Once a pathogenic organism has been isolated, it can be further characterised by its morphology, growth patterns (such as aerobic or anaerobic growth), patterns of hemolysis, and staining.[177]
As with bacterial classification, identification of bacteria is increasingly using molecular methods,[178] and mass spectroscopy.[179] Most bacteria have not been characterised and there are may species that cannot be grown in the laboratory.[180] Diagnostics using DNA-based tools, such as polymerase chain reaction, are increasingly popular due to their specificity and speed, compared to culture-based methods.[181] These methods also allow the detection and identification of "viable but nonculturable" cells that are metabolically active but non-dividing.[182] However, even using these improved methods, the total number of bacterial species is not known and cannot even be estimated with any certainty. Following present classification, there are a little less than 9,300 known species of prokaryotes, which includes bacteria and archaea;[183] but attempts to estimate the true number of bacterial diversity have ranged from 107 to 109 total species—and even these diverse estimates may be off by many orders of magnitude.[184][185]
Phyla
Valid phyla
The following phyla have been validly published according to the Bacteriological Code:[186]
- Acidobacteriota
- Actinomycetota
- Aquificota
- Armatimonadota
- Atribacterota
- Bacillota
- Bacteroidota
- Balneolota
- Bdellovibrionota
- Caldisericota
- Calditrichota
- Campylobacterota
- Chlamydiota
- Chlorobiota
- Chloroflexota
- Chrysiogenota
- Coprothermobacterota
- Deferribacterota
- Deinococcota
- Dictyoglomota
- Elusimicrobiota
- Fibrobacterota
- Fusobacteriota
- Gemmatimonadota
- Ignavibacteriota
- Lentisphaerota
- Mycoplasmatota
- Myxococcota
- Nitrospinota
- Nitrospirota
- Planctomycetota
- Pseudomonadota
- Rhodothermota
- Spirochaetota
- Synergistota
- Thermodesulfobacteriota
- Thermomicrobiota
- Thermotogota
- Verrucomicrobiota
Provisional phyla
The following phyla have been proposed, but have not been validly published according to the Bacteriological Code (including those that have candidatus status):[187][188][189][190]
- "Candidatus Abawacabacteria"
- "Abditibacteriota"
- "Candidatus Absconditabacteria"
- "Candidatus Acetothermia"
- "Candidatus Adlerbacteria"
- "Candidatus Aerophobetes"
- "Candidatus Amesbacteria"
- "Candidatus Aminicenantes"
- "Candidatus Andersenbacteria"
- "Candidatus Azambacteria"
- "Candidatus Beckwithbacteria"
- "Candidatus Berkelbacteria"
- "Candidatus Binatota"
- "Candidatus Bipolaricaulota"
- "Candidatus Blackallbacteria"
- "Candidatus Blackburnbacteria"
- "Candidatus Brennerbacteria"
- "Candidatus Brownbacteria"
- "Candidatus Buchananbacteria"
- "Candidatus Caldatribacteriota"
- "Candidatus Calescamantes"
- "Candidatus Campbellbacteria"
- "Candidatus Chisholmbacteria"
- "Candidatus Cloacimonetes"
- "Candidatus Coatesbacteria"
- "Candidatus Collierbacteria"
- "Candidatus Colwellbacteria"
- "Candidatus Cryosericota"
- "Candidatus Curtissbacteria"
- "Cyanobacteria"
- "Candidatus Dadabacteria"
- "Candidatus Daviesbacteria"
- "Candidatus Delongbacteria"
- "Candidatus Delphibacteria"
- "Candidatus Dependentiae"
- "Candidatus Desantisbacteria"
- "Candidatus Dojkabacteria"
- "Candidatus Dormibacteraeota"
- "Candidatus Doudnabacteria"
- "Candidatus Edwardsbacteria"
- "Candidatus Eisenbacteria"
- "Candidatus Elulimicrobiota"
- "Candidatus Eremiobacterota"
- "Candidatus Falkowbacteria"
- "Candidatus Fermentibacteria"
- "Candidatus Fertabacteria"
- "Candidatus Fervidibacteria"
- "Candidatus Firestonebacteria"
- "Candidatus Fischerbacteria"
- "Candidatus Fraserbacteria"
- "Candidatus Genascibacteria"
- "Candidatus Giovannonibacteria"
- "Candidatus Glassbacteria"
- "Candidatus Goldbacteria"
- "Candidatus Gottesmanbacteria"
- "Candidatus Gracilibacteria"
- "Candidatus Gribaldobacteria"
- "Candidatus Handelsmanbacteria"
- "Candidatus Harrisonbacteria"
- "Candidatus Howlettbacteria"
- "Candidatus Hugbacteria"
- "Candidatus Hydrogenedentes"
- "Candidatus Hydrothermae"
- "Candidatus Hydrothermota"
- "Candidatus Jacksonbacteria"
- "Candidatus Jorgensenbacteria"
- "Candidatus Kaiserbacteria"
- "Candidatus Kapabacteria"
- "Candidatus Katanobacteria"
- "Candidatus Kerfeldbacteria"
- "Candidatus Komeilibacteria"
- "Candidatus Krumholzibacteriota"
- "Candidatus Kryptonia"
- "Candidatus Kuenenbacteria"
- "Candidatus Lambdaproteobacteria"
- "Candidatus Latescibacteria"
- "Candidatus Levybacteria"
- "Candidatus Lindowbacteria"
- "Candidatus Liptonbacteria"
- "Candidatus Lloydbacteria"
- "Candidatus Magasanikbacteria"
- "Candidatus Margulisbacteria"
- "Candidatus Marinimicrobia"
- "Candidatus Mcinerneyibacteriota"
- "Candidatus Melainabacteria"
- "Candidatus Microgenomates"
- "Candidatus Modulibacteria"
- "Candidatus Moisslbacteria"
- "Candidatus Montesolbacteria"
- "Candidatus Moranbacteria"
- "Candidatus Muirbacteria"
- "Candidatus Muproteobacteria"
- "Candidatus Nealsonbacteria"
- "Candidatus Niyogibacteria"
- "Candidatus Nomurabacteria"
- "Candidatus Omnitrophica"
- "Candidatus Pacebacteria"
- "Candidatus Parcubacteria"
- "Candidatus Parcunitrobacteria"
- "Candidatus Peregrinibacteria"
- "Candidatus Poribacteria"
- "Candidatus Portnoybacteria"
- "Candidatus Pyropristinus"
- "Candidatus Ratteibacteria"
- "Candidatus Raymondbacteria"
- "Candidatus Riflebacteria"
- "Candidatus Roizmanbacteria"
- "Candidatus Rokubacteria"
- "Candidatus Ryanbacteria"
- "Candidatus Saccharibacteria"
- "Candidatus Saganbacteria"
- "Candidatus Schekmanbacteria"
- "Candidatus Shapirobacteria"
- "Candidatus Spechtbacteria"
- "Candidatus Stahlbacteria"
- "Candidatus Staskawiczbacteria"
- "Candidatus Sumerlaeota"
- "Candidatus Sungbacteria"
- "Candidatus Tagabacteria"
- "Candidatus Taylorbacteria"
- "Candidatus Tectomicrobia"
- "Candidatus Terrybacteria"
- "Candidatus Teskebacteria"
- "Candidatus Tianyabacteria"
- "Candidatus Torokbacteria"
- "Candidatus Uhrbacteria"
- "Candidatus Veblenbacteria"
- "Candidatus Vogelbacteria"
- "Candidatus Wallbacteria"
- "Candidatus Wildermuthbacteria"
- "Candidatus Wirthbacteria"
- "Candidatus Woesebacteria"
- "Candidatus Wolfebacteria"
- "Candidatus Woykebacteria"
- "Candidatus Yanofskybacteria"
- "Candidatus Yonathbacteria"
- "Candidatus Zambryskibacteria"
- "Candidatus Zixibacteria"
Genera incertae sedis
The following bacteria genera have not been assigned to a phylum, class, or order:
- "Fermentobadaceae" Haiying 1995
- "Guhaiyingella" Haiying 1995
- Not assigned to a family:
- "Candidatus Aegiribacteria" Hamilton et al. 2016
- Archaeoscillatoriopsis Schopf 1993
- "Eoleptonema" Awramik et al. 1983
- "Candidatus Epulonipiscium" corrig. Montgomery and Pollak 1988
- "Candidatus Ovibacter" corrig. Fenchel and Thar 2004
- "Primaevifilum" Schopf 1983
- "Rappaport" Waldman Ben-Asher et al. 2017
Interactions with other organisms
Despite their apparent simplicity, bacteria can form complex associations with other organisms. These symbiotic associations can be divided into parasitism, mutualism and commensalism.[192]
Commensals
The word "commensalism" is derived from the word "commensal", meaning "eating at the same table"[193] and all plants and animals are colonised by commensal bacteria. In humans and other animals millions of them live on the skin, the airways, the gut and other orifices.[194][195] Referred to as "normal flora",[196] or "commensals",[197] these bacteria usually cause no harm but may occasionally invade other sites of the body and cause infection. Escherichia coli is a commensal in the human gut but can cause urinary tract infections.[198] Similarly, streptoccoci, which are part of the normal flora of the human mouth, can cause heart disease.[199]
Predators
Some species of bacteria kill and then consume other microorganisms, these species are called predatory bacteria.[200] These include organisms such as Myxococcus xanthus, which forms swarms of cells that kill and digest any bacteria they encounter.[201] Other bacterial predators either attach to their prey in order to digest them and absorb nutrients or invade another cell and multiply inside the cytosol.[202] These predatory bacteria are thought to have evolved from saprophages that consumed dead microorganisms, through adaptations that allowed them to entrap and kill other organisms.[203]
Mutualists
Certain bacteria form close spatial associations that are essential for their survival. One such mutualistic association, called interspecies hydrogen transfer, occurs between clusters of anaerobic bacteria that consume organic acids, such as butyric acid or propionic acid, and produce hydrogen, and methanogenic archaea that consume hydrogen.[204] The bacteria in this association are unable to consume the organic acids as this reaction produces hydrogen that accumulates in their surroundings. Only the intimate association with the hydrogen-consuming archaea keeps the hydrogen concentration low enough to allow the bacteria to grow.[205]
In soil, microorganisms that reside in the rhizosphere (a zone that includes the root surface and the soil that adheres to the root after gentle shaking) carry out nitrogen fixation, converting nitrogen gas to nitrogenous compounds.[206] This serves to provide an easily absorbable form of nitrogen for many plants, which cannot fix nitrogen themselves. Many other bacteria are found as symbionts in humans and other organisms. For example, the presence of over 1,000 bacterial species in the normal human gut flora of the intestines can contribute to gut immunity, synthesise vitamins, such as folic acid, vitamin K and biotin, convert sugars to lactic acid (see Lactobacillus), as well as fermenting complex undigestible carbohydrates.[207][208][209] The presence of this gut flora also inhibits the growth of potentially pathogenic bacteria (usually through competitive exclusion) and these beneficial bacteria are consequently sold as probiotic dietary supplements.[210]
Nearly all animal life is dependent on bacteria for survival as only bacteria and some archaea possess the genes and enzymes necessary to synthesize vitamin B12, also known as cobalamin, and provide it through the food chain. Vitamin B12 is a water-soluble vitamin that is involved in the metabolism of every cell of the human body. It is a cofactor in DNA synthesis, and in both fatty acid and amino acid metabolism. It is particularly important in the normal functioning of the nervous system via its role in the synthesis of myelin.[211]
Pathogens
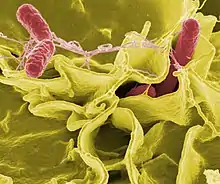
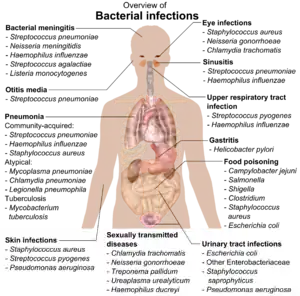
The body is continually exposed to many species of bacteria, including beneficial commensals, which grow on the skin and mucous membranes, and saprophytes, which grow mainly in the soil and in decaying matter. The blood and tissue fluids contain nutrients sufficient to sustain the growth of many bacteria. The body has defence mechanisms that enable it to resist microbial invasion of its tissues and give it a natural immunity or innate resistance against many microorganisms.[212] Unlike some viruses, bacteria evolve relatively slowly so many bacterial diseases also occur in other animals.[213]
If bacteria form a parasitic association with other organisms, they are classed as pathogens.[214] Pathogenic bacteria are a major cause of human death and disease and cause infections such as tetanus (caused by Clostridium tetani), typhoid fever, diphtheria, syphilis, cholera, foodborne illness, leprosy (caused by Mycobacterium leprae) and tuberculosis (caused by Mycobacterium tuberculosis).[215] A pathogenic cause for a known medical disease may only be discovered many years later, as was the case with Helicobacter pylori and peptic ulcer disease.[216] Bacterial diseases are also important in agriculture, with bacteria causing leaf spot, fire blight and wilts in plants, as well as Johne's disease, mastitis, salmonella and anthrax in farm animals.[217]
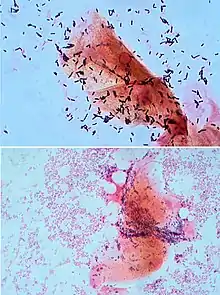
Each species of pathogen has a characteristic spectrum of interactions with its human hosts. Some organisms, such as Staphylococcus or Streptococcus, can cause skin infections, pneumonia, meningitis and sepsis, a systemic inflammatory response producing shock, massive vasodilation and death.[218] Yet these organisms are also part of the normal human flora and usually exist on the skin or in the nose without causing any disease at all. Other organisms invariably cause disease in humans, such as Rickettsia, which are obligate intracellular parasites able to grow and reproduce only within the cells of other organisms. One species of Rickettsia causes typhus, while another causes Rocky Mountain spotted fever. Chlamydia, another phylum of obligate intracellular parasites, contains species that can cause pneumonia or urinary tract infection and may be involved in coronary heart disease.[219] Some species, such as Pseudomonas aeruginosa, Burkholderia cenocepacia, and Mycobacterium avium, are opportunistic pathogens and cause disease mainly in people who are immunosuppressed or have cystic fibrosis.[220][221] Some bacteria produce toxins, which cause diseases.[222] These are endotoxins, which come from broken bacterial cells, and exotoxins, which are produced by bacteria and released into the environment.[223] The bacterium Clostridium botulinum for example, produces a powerful exotoxin that cause respiratory paralysis, and Salmonellae produce an endotoxin that causes gastroenteritis.[223] Some exotoxins can be converted to toxoids, which are used as vaccines to prevent the disease.[224]
Bacterial infections may be treated with antibiotics, which are classified as bacteriocidal if they kill bacteria or bacteriostatic if they just prevent bacterial growth. There are many types of antibiotics, and each class inhibits a process that is different in the pathogen from that found in the host. An example of how antibiotics produce selective toxicity are chloramphenicol and puromycin, which inhibit the bacterial ribosome, but not the structurally different eukaryotic ribosome.[225] Antibiotics are used both in treating human disease and in intensive farming to promote animal growth, where they may be contributing to the rapid development of antibiotic resistance in bacterial populations.[226] Infections can be prevented by antiseptic measures such as sterilising the skin prior to piercing it with the needle of a syringe, and by proper care of indwelling catheters. Surgical and dental instruments are also sterilised to prevent contamination by bacteria. Disinfectants such as bleach are used to kill bacteria or other pathogens on surfaces to prevent contamination and further reduce the risk of infection.[227]
Significance in technology and industry
Bacteria, often lactic acid bacteria, such as Lactobacillus species and Lactococcus species, in combination with yeasts and moulds, have been used for thousands of years in the preparation of fermented foods, such as cheese, pickles, soy sauce, sauerkraut, vinegar, wine and yogurt.[228][229]
The ability of bacteria to degrade a variety of organic compounds is remarkable and has been used in waste processing and bioremediation. Bacteria capable of digesting the hydrocarbons in petroleum are often used to clean up oil spills.[230] Fertiliser was added to some of the beaches in Prince William Sound in an attempt to promote the growth of these naturally occurring bacteria after the 1989 Exxon Valdez oil spill. These efforts were effective on beaches that were not too thickly covered in oil. Bacteria are also used for the bioremediation of industrial toxic wastes.[231] In the chemical industry, bacteria are most important in the production of enantiomerically pure chemicals for use as pharmaceuticals or agrichemicals.[232]
Bacteria can also be used in the place of pesticides in the biological pest control. This commonly involves Bacillus thuringiensis (also called BT), a Gram-positive, soil dwelling bacterium. Subspecies of this bacteria are used as a Lepidopteran-specific insecticides under trade names such as Dipel and Thuricide.[233] Because of their specificity, these pesticides are regarded as environmentally friendly, with little or no effect on humans, wildlife, pollinators and most other beneficial insects.[234][235]
Because of their ability to quickly grow and the relative ease with which they can be manipulated, bacteria are the workhorses for the fields of molecular biology, genetics and biochemistry. By making mutations in bacterial DNA and examining the resulting phenotypes, scientists can determine the function of genes, enzymes and metabolic pathways in bacteria, then apply this knowledge to more complex organisms.[236] This aim of understanding the biochemistry of a cell reaches its most complex expression in the synthesis of huge amounts of enzyme kinetic and gene expression data into mathematical models of entire organisms. This is achievable in some well-studied bacteria, with models of Escherichia coli metabolism now being produced and tested.[237][238] This understanding of bacterial metabolism and genetics allows the use of biotechnology to bioengineer bacteria for the production of therapeutic proteins, such as insulin, growth factors, or antibodies.[239][240]
Because of their importance for research in general, samples of bacterial strains are isolated and preserved in Biological Resource Centers. This ensures the availability of the strain to scientists worldwide.[241]
History of bacteriology
._Natuurkundige_te_Delft_Rijksmuseum_SK-A-957.jpeg.webp)
Bacteria were first observed by the Dutch microscopist Antonie van Leeuwenhoek in 1676, using a single-lens microscope of his own design. He then published his observations in a series of letters to the Royal Society of London.[242] Bacteria were Leeuwenhoek's most remarkable microscopic discovery. They were just at the limit of what his simple lenses could make out and, in one of the most striking hiatuses in the history of science, no one else would see them again for over a century.[243] His observations had also included protozoans which he called animalcules, and his findings were looked at again in the light of the more recent findings of cell theory.[244]
Christian Gottfried Ehrenberg introduced the word "bacterium" in 1828.[245] In fact, his Bacterium was a genus that contained non-spore-forming rod-shaped bacteria,[246] as opposed to Bacillus, a genus of spore-forming rod-shaped bacteria defined by Ehrenberg in 1835.[247]
Louis Pasteur demonstrated in 1859 that the growth of microorganisms causes the fermentation process, and that this growth is not due to spontaneous generation (yeasts and molds, commonly associated with fermentation, are not bacteria, but rather fungi). Along with his contemporary Robert Koch, Pasteur was an early advocate of the germ theory of disease.[248] Before them, Ignaz Semmelweis and Joseph Lister had realised the importance of sanitized hands in medical work. Semmelweis ideas was rejected and his book on the topic condemned by the medical community, but after Lister doctors started sanitizing their hands in the 1870s. While Semmelweis who started with rules about handwashing in his hospital in the 1840s predated the spread of the ideas about germs themselves and attributed diseases to "decomposing animal organic matter", Lister was active later.[249]
Robert Koch, a pioneer in medical microbiology, worked on cholera, anthrax and tuberculosis. In his research into tuberculosis Koch finally proved the germ theory, for which he received a Nobel Prize in 1905.[250] In Koch's postulates, he set out criteria to test if an organism is the cause of a disease, and these postulates are still used today.[251]
Ferdinand Cohn is said to be a founder of bacteriology, studying bacteria from 1870. Cohn was the first to classify bacteria based on their morphology.[252][253]
Though it was known in the nineteenth century that bacteria are the cause of many diseases, no effective antibacterial treatments were available.[254] In 1910, Paul Ehrlich developed the first antibiotic, by changing dyes that selectively stained Treponema pallidum—the spirochaete that causes syphilis—into compounds that selectively killed the pathogen.[255] Ehrlich had been awarded a 1908 Nobel Prize for his work on immunology, and pioneered the use of stains to detect and identify bacteria, with his work being the basis of the Gram stain and the Ziehl–Neelsen stain.[256]
A major step forward in the study of bacteria came in 1977 when Carl Woese recognised that archaea have a separate line of evolutionary descent from bacteria.[257] This new phylogenetic taxonomy depended on the sequencing of 16S ribosomal RNA, and divided prokaryotes into two evolutionary domains, as part of the three-domain system.[3]
See also
- Genetically modified bacteria
- Marine prokaryotes
References
- "31. Ancient Life: Apex Chert Microfossils". www.lpi.usra.edu. Retrieved 12 March 2022.
- McCutcheon, JP (6 October 2021). "The Genomics and Cell Biology of Host-Beneficial Intracellular Infections". Annual Review of Cell and Developmental Biology. 37 (1): 115–142. doi:10.1146/annurev-cellbio-120219-024122. ISSN 1081-0706. PMID 34242059. S2CID 235786110. Archived from the original on 7 July 2022. Retrieved 19 August 2022.
- Hall 2008, p. 145.
- βακτήριον. Liddell, Henry George; Scott, Robert; A Greek–English Lexicon at the Perseus Project.
- βακτηρία in Liddell and Scott.
- Harper, Douglas. "bacteria". Online Etymology Dictionary.
- Krasner 2014, p. 74.
- Woese CR, Kandler O, Wheelis ML (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–79. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Hall 2008, p. 84.
- Godoy-Vitorino F (July 2019). "Human microbial ecology and the rising new medicine". Annals of Translational Medicine. 7 (14): 342. doi:10.21037/atm.2019.06.56. PMC 6694241. PMID 31475212.
- Schopf JW (July 1994). "Disparate rates, differing fates: tempo and mode of evolution changed from the Precambrian to the Phanerozoic". Proceedings of the National Academy of Sciences of the United States of America. 91 (15): 6735–42. Bibcode:1994PNAS...91.6735S. doi:10.1073/pnas.91.15.6735. PMC 44277. PMID 8041691.
- DeLong EF, Pace NR (August 2001). "Environmental diversity of bacteria and archaea". Systematic Biology. 50 (4): 470–78. CiteSeerX 10.1.1.321.8828. doi:10.1080/106351501750435040. PMID 12116647.
- Brown JR, Doolittle WF (December 1997). "Archaea and the prokaryote-to-eukaryote transition". Microbiology and Molecular Biology Reviews. 61 (4): 456–502. doi:10.1128/mmbr.61.4.456-502.1997. PMC 232621. PMID 9409149.
- Daum B, Gold V (June 2018). "Twitch or swim: towards the understanding of prokaryotic motion based on the type IV pilus blueprint". Biological Chemistry. 399 (7): 799–808. doi:10.1515/hsz-2018-0157. hdl:10871/33366. PMID 29894297. S2CID 48352675.
- Di Giulio M (December 2003). "The universal ancestor and the ancestor of bacteria were hyperthermophiles". Journal of Molecular Evolution. 57 (6): 721–30. Bibcode:2003JMolE..57..721D. doi:10.1007/s00239-003-2522-6. PMID 14745541. S2CID 7041325.
- Battistuzzi FU, Feijao A, Hedges SB (November 2004). "A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land". BMC Evolutionary Biology. 4: 44. doi:10.1186/1471-2148-4-44. PMC 533871. PMID 15535883.
- Homann, Martin; et al. (23 July 2018). "Microbial life and biogeochemical cycling on land 3,220 million years ago" (PDF). Nature Geoscience. 11 (9): 665–671. Bibcode:2018NatGe..11..665H. doi:10.1038/s41561-018-0190-9. S2CID 134935568.
- Gabaldón, T (8 October 2021). "Origin and Early Evolution of the Eukaryotic Cell". Annual Review of Microbiology. 75 (1): 631–647. doi:10.1146/annurev-micro-090817-062213. ISSN 0066-4227. PMID 34343017. S2CID 236916203. Archived from the original on 19 August 2022. Retrieved 19 August 2022.
- Callier, Viviane (8 June 2022). "Mitochondria and the origin of eukaryotes". Knowable Magazine. doi:10.1146/knowable-060822-2. Retrieved 19 August 2022.
- Poole AM, Penny D (January 2007). "Evaluating hypotheses for the origin of eukaryotes". BioEssays. 29 (1): 74–84. doi:10.1002/bies.20516. PMID 17187354.
- Dyall SD, Brown MT, Johnson PJ (April 2004). "Ancient invasions: from endosymbionts to organelles". Science. 304 (5668): 253–57. Bibcode:2004Sci...304..253D. doi:10.1126/science.1094884. PMID 15073369. S2CID 19424594.
- Stephens TG, Gabr A, Calatrava V, Grossman AR, Bhattacharya D (May 2021). "Why is primary endosymbiosis so rare?". The New Phytologist. 231 (5): 1693–1699. doi:10.1111/nph.17478. PMC 8711089. PMID 34018613.
- Baker-Austin C, Dopson M (April 2007). "Life in acid: pH homeostasis in acidophiles". Trends in Microbiology. 15 (4): 165–71. doi:10.1016/j.tim.2007.02.005. PMID 17331729.
- Jeong SW, Choi YJ (October 2020). "Extremophilic Microorganisms for the Treatment of Toxic Pollutants in the Environment". Molecules (Basel, Switzerland). 25 (21): 4916. doi:10.3390/molecules25214916. PMC 7660605. PMID 33114255.
- Flemming HC, Wuertz S (April 2019). "Bacteria and archaea on Earth and their abundance in biofilms". Nature Reviews. Microbiology. 17 (4): 247–260. doi:10.1038/s41579-019-0158-9. PMID 30760902. S2CID 61155774.
- Bar-On YM, Phillips R, Milo R (June 2018). "The biomass distribution on Earth". Proceedings of the National Academy of Sciences of the United States of America. 115 (25): 6506–11. Bibcode:2018PNAS..115.6506B. doi:10.1073/pnas.1711842115. PMC 6016768. PMID 29784790.
- Wheelis 2008, p. 362.
- Kushkevych I, Procházka J, Gajdács M, Rittmann SK, Vítězová M (June 2021). "Molecular Physiology of Anaerobic Phototrophic Purple and Green Sulfur Bacteria". International Journal of Molecular Sciences. 22 (12): 6398. doi:10.3390/ijms22126398. PMC 8232776. PMID 34203823.
- Wheelis 2008, p. 6.
- Pommerville 2014, p. 3–6.
- Krasner 2014, p. 38.
- Pommerville 2014, p. 134.
- Schulz HN, Jorgensen BB (2001). "Big bacteria". Annual Review of Microbiology. 55: 105–137. doi:10.1146/annurev.micro.55.1.105. PMID 11544351. S2CID 18168018.
- Williams C (2011). "Who are you calling simple?". New Scientist. 211 (2821): 38–41. doi:10.1016/S0262-4079(11)61709-0.
- Volland JM, Gonzalez-Rizzo S, Gros O, Tyml T, Ivanova N, Schulz F, Goudeau D, Elisabeth NH, Nath N, Udwary D, Malmstrom RR (18 February 2022). "A centimeter-long bacterium with DNA compartmentalized in membrane-bound organelles". bioRxiv (preprint). doi:10.1101/2022.02.16.480423. S2CID 246975579.
- Sanderson K (June 2022). "Largest bacterium ever found is surprisingly complex". Nature. doi:10.1038/d41586-022-01757-1. PMID 35750919. S2CID 250022076.
- Robertson J, Gomersall M, Gill P (November 1975). "Mycoplasma hominis: growth, reproduction, and isolation of small viable cells". Journal of Bacteriology. 124 (2): 1007–1018. doi:10.1128/JB.124.2.1007-1018.1975. PMC 235991. PMID 1102522.
- Velimirov B (2001). "Nanobacteria, Ultramicrobacteria and Starvation Forms: A Search for the Smallest Metabolizing Bacterium". Microbes and Environments. 16 (2): 67–77. doi:10.1264/jsme2.2001.67.
- Dusenbery, David B (2009). Living at Micro Scale, pp. 20–25. Harvard University Press, Cambridge, Massachusetts ISBN 978-0-674-03116-6.
- Yang DC, Blair KM, Salama NR (March 2016). "Staying in Shape: the Impact of Cell Shape on Bacterial Survival in Diverse Environments". Microbiology and Molecular Biology Reviews. 80 (1): 187–203. doi:10.1128/MMBR.00031-15. PMC 4771367. PMID 26864431.
- Cabeen MT, Jacobs-Wagner C (August 2005). "Bacterial cell shape". Nature Reviews. Microbiology. 3 (8): 601–10. doi:10.1038/nrmicro1205. PMID 16012516. S2CID 23938989.
- Young KD (September 2006). "The selective value of bacterial shape". Microbiology and Molecular Biology Reviews. 70 (3): 660–703. doi:10.1128/MMBR.00001-06. PMC 1594593. PMID 16959965.
- Crawford 2007, p. xi.
- Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, van Wezel GP (February 2014). "Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies". Nature Reviews. Microbiology. 12 (2): 115–24. doi:10.1038/nrmicro3178. hdl:11370/0db66a9c-72ef-4e11-a75d-9d1e5827573d. PMID 24384602. S2CID 20154495.
- Shimkets LJ (1999). "Intercellular signaling during fruiting-body development of Myxococcus xanthus". Annual Review of Microbiology. 53: 525–49. doi:10.1146/annurev.micro.53.1.525. PMID 10547700.
- Kaiser D (2004). "Signaling in myxobacteria". Annual Review of Microbiology. 58: 75–98. doi:10.1146/annurev.micro.58.030603.123620. PMID 15487930.
- Wheelis 2008, p. 75.
- Mandal A, Dutta A, Das R, Mukherjee J (June 2021). "Role of intertidal microbial communities in carbon dioxide sequestration and pollutant removal: A review". Marine Pollution Bulletin. 170: 112626. doi:10.1016/j.marpolbul.2021.112626. PMID 34153859.
- Donlan RM (September 2002). "Biofilms: microbial life on surfaces". Emerging Infectious Diseases. 8 (9): 881–90. doi:10.3201/eid0809.020063. PMC 2732559. PMID 12194761.
- Branda SS, Vik S, Friedman L, Kolter R (January 2005). "Biofilms: the matrix revisited". Trends in Microbiology. 13 (1): 20–26. doi:10.1016/j.tim.2004.11.006. PMID 15639628.
- Davey ME, O'toole GA (December 2000). "Microbial biofilms: from ecology to molecular genetics". Microbiology and Molecular Biology Reviews. 64 (4): 847–67. doi:10.1128/MMBR.64.4.847-867.2000. PMC 99016. PMID 11104821.
- Donlan RM, Costerton JW (April 2002). "Biofilms: survival mechanisms of clinically relevant microorganisms". Clinical Microbiology Reviews. 15 (2): 167–93. doi:10.1128/CMR.15.2.167-193.2002. PMC 118068. PMID 11932229.
- Slonczewski JL, Foster JW (2013). Microbiology : an Evolving Science (Third ed.). New York: W W Norton. p. 82. ISBN 978-0-393-12367-8.
- Feijoo-Siota L, Rama JL, Sánchez-Pérez A, Villa TG (July 2017). "Considerations on bacterial nucleoids". Applied Microbiology and Biotechnology. 101 (14): 5591–602. doi:10.1007/s00253-017-8381-7. PMID 28664324. S2CID 10173266.
- Bobik TA (May 2006). "Polyhedral organelles compartmenting bacterial metabolic processes". Applied Microbiology and Biotechnology. 70 (5): 517–25. doi:10.1007/s00253-005-0295-0. PMID 16525780. S2CID 8202321.
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (September 2008). "Protein-based organelles in bacteria: carboxysomes and related microcompartments". Nature Reviews. Microbiology. 6 (9): 681–91. doi:10.1038/nrmicro1913. PMID 18679172. S2CID 22666203.
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (August 2005). "Protein structures forming the shell of primitive bacterial organelles". Science. 309 (5736): 936–38. Bibcode:2005Sci...309..936K. CiteSeerX 10.1.1.1026.896. doi:10.1126/science.1113397. PMID 16081736. S2CID 24561197.
- Gitai Z (March 2005). "The new bacterial cell biology: moving parts and subcellular architecture". Cell. 120 (5): 577–86. doi:10.1016/j.cell.2005.02.026. PMID 15766522. S2CID 8894304.
- Shih YL, Rothfield L (September 2006). "The bacterial cytoskeleton". Microbiology and Molecular Biology Reviews. 70 (3): 729–54. doi:10.1128/MMBR.00017-06. PMC 1594594. PMID 16959967.
- Norris V, den Blaauwen T, Cabin-Flaman A, Doi RH, Harshey R, Janniere L, Jimenez-Sanchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Saier M, Skarstad K (March 2007). "Functional taxonomy of bacterial hyperstructures". Microbiology and Molecular Biology Reviews. 71 (1): 230–53. doi:10.1128/MMBR.00035-06. PMC 1847379. PMID 17347523.
- Pommerville 2014, pp. 120–121.
- Bryant DA, Frigaard NU (November 2006). "Prokaryotic photosynthesis and phototrophy illuminated". Trends in Microbiology. 14 (11): 488–96. doi:10.1016/j.tim.2006.09.001. PMID 16997562.
- Psencík J, Ikonen TP, Laurinmäki P, Merckel MC, Butcher SJ, Serimaa RE, Tuma R (August 2004). "Lamellar organization of pigments in chlorosomes, the light harvesting complexes of green photosynthetic bacteria". Biophysical Journal. 87 (2): 1165–72. Bibcode:2004BpJ....87.1165P. doi:10.1529/biophysj.104.040956. PMC 1304455. PMID 15298919.
- Thanbichler M, Wang SC, Shapiro L (October 2005). "The bacterial nucleoid: a highly organized and dynamic structure". Journal of Cellular Biochemistry. 96 (3): 506–21. doi:10.1002/jcb.20519. PMID 15988757. S2CID 25355087.
- Poehlsgaard J, Douthwaite S (November 2005). "The bacterial ribosome as a target for antibiotics". Nature Reviews. Microbiology. 3 (11): 870–81. doi:10.1038/nrmicro1265. PMID 16261170. S2CID 7521924.
- Yeo M, Chater K (March 2005). "The interplay of glycogen metabolism and differentiation provides an insight into the developmental biology of Streptomyces coelicolor". Microbiology. 151 (Pt 3): 855–61. doi:10.1099/mic.0.27428-0. PMID 15758231. Archived from the original on 29 September 2007.
- Shiba T, Tsutsumi K, Ishige K, Noguchi T (March 2000). "Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications". Biochemistry. Biokhimiia. 65 (3): 315–23. PMID 10739474. Archived from the original on 25 September 2006.
- Brune DC (June 1995). "Isolation and characterization of sulfur globule proteins from Chromatium vinosum and Thiocapsa roseopersicina". Archives of Microbiology. 163 (6): 391–99. doi:10.1007/BF00272127. PMID 7575095. S2CID 22279133.
- Kadouri D, Jurkevitch E, Okon Y, Castro-Sowinski S (2005). "Ecological and agricultural significance of bacterial polyhydroxyalkanoates". Critical Reviews in Microbiology. 31 (2): 55–67. doi:10.1080/10408410590899228. PMID 15986831. S2CID 4098268.
- Walsby AE (March 1994). "Gas vesicles". Microbiological Reviews. 58 (1): 94–144. doi:10.1128/MMBR.58.1.94-144.1994. PMC 372955. PMID 8177173.
- van Heijenoort J (March 2001). "Formation of the glycan chains in the synthesis of bacterial peptidoglycan". Glycobiology. 11 (3): 25R–36R. doi:10.1093/glycob/11.3.25R. PMID 11320055. S2CID 46066256.
- Koch AL (October 2003). "Bacterial wall as target for attack: past, present, and future research". Clinical Microbiology Reviews. 16 (4): 673–87. doi:10.1128/CMR.16.4.673-687.2003. PMC 207114. PMID 14557293.
- Gram HC (1884). "Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten". Fortschr. Med. 2: 185–89.
- Hugenholtz P (2002). "Exploring prokaryotic diversity in the genomic era". Genome Biology. 3 (2): REVIEWS0003. doi:10.1186/gb-2002-3-2-reviews0003. PMC 139013. PMID 11864374.
- Walsh FM, Amyes SG (October 2004). "Microbiology and drug resistance mechanisms of fully resistant pathogens" (PDF). Current Opinion in Microbiology. 7 (5): 439–44. doi:10.1016/j.mib.2004.08.007. PMID 15451497.
- Alderwick LJ, Harrison J, Lloyd GS, Birch HL (March 2015). "The Mycobacterial Cell Wall – Peptidoglycan and Arabinogalactan". Cold Spring Harbor Perspectives in Medicine. 5 (8): a021113. doi:10.1101/cshperspect.a021113. PMC 4526729. PMID 25818664.
- Fagan RP, Fairweather NF (March 2014). "Biogenesis and functions of bacterial S-layers" (PDF). Nature Reviews. Microbiology. 12 (3): 211–22. doi:10.1038/nrmicro3213. PMID 24509785. S2CID 24112697.
- Thompson SA (December 2002). "Campylobacter surface-layers (S-layers) and immune evasion". Annals of Periodontology. 7 (1): 43–53. doi:10.1902/annals.2002.7.1.43. PMC 2763180. PMID 16013216.
- Beveridge TJ, Pouwels PH, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer EM, Schocher I, Sleytr UB, Morelli L, Callegari ML, Nomellini JF, Bingle WH, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Béguin P, Ohayon H, Gounon P, Matuschek M, Koval SF (June 1997). "Functions of S-layers". FEMS Microbiology Reviews. 20 (1–2): 99–149. doi:10.1016/S0168-6445(97)00043-0. PMID 9276929.
- Kojima S, Blair DF (2004). The bacterial flagellar motor: structure and function of a complex molecular machine. International Review of Cytology. Vol. 233. pp. 93–134. doi:10.1016/S0074-7696(04)33003-2. ISBN 978-0-12-364637-8. PMID 15037363.
- Wheelis 2008, p. 76.
- Cheng RA, Wiedmann M (2020). "Recent Advances in Our Understanding of the Diversity and Roles of Chaperone-Usher Fimbriae in Facilitating Salmonella Host and Tissue Tropism". Frontiers in Cellular and Infection Microbiology. 10: 628043. doi:10.3389/fcimb.2020.628043. PMC 7886704. PMID 33614531.
- Silverman PM (February 1997). "Towards a structural biology of bacterial conjugation". Molecular Microbiology. 23 (3): 423–29. doi:10.1046/j.1365-2958.1997.2411604.x. PMID 9044277. S2CID 24126399.
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G (June 2015). "Secretion systems in Gram-negative bacteria: structural and mechanistic insights". Nature Reviews. Microbiology. 13 (6): 343–59. doi:10.1038/nrmicro3456. PMID 25978706. S2CID 8664247.
- Luong P, Dube DH (July 2021). "Dismantling the bacterial glycocalyx: Chemical tools to probe, perturb, and image bacterial glycans". Bioorganic & Medicinal Chemistry. 42: 116268. doi:10.1016/j.bmc.2021.116268. ISSN 0968-0896. PMC 8276522. PMID 34130219.
- Stokes RW, Norris-Jones R, Brooks DE, Beveridge TJ, Doxsee D, Thorson LM (October 2004). "The glycan-rich outer layer of the cell wall of Mycobacterium tuberculosis acts as an antiphagocytic capsule limiting the association of the bacterium with macrophages". Infection and Immunity. 72 (10): 5676–86. doi:10.1128/IAI.72.10.5676-5686.2004. PMC 517526. PMID 15385466.
- Kalscheuer R, Palacios A, Anso I, Cifuente J, Anguita J, Jacobs WR, Guerin ME, Prados-Rosales R (July 2019). "The Mycobacterium tuberculosis capsule: a cell structure with key implications in pathogenesis". The Biochemical Journal. 476 (14): 1995–2016. doi:10.1042/BCJ20190324. PMC 6698057. PMID 31320388.
- Jernigan JA, Stephens DS, Ashford DA, Omenaca C, Topiel MS, Galbraith M, Tapper M, Fisk TL, Zaki S, Popovic T, Meyer RF, Quinn CP, Harper SA, Fridkin SK, Sejvar JJ, Shepard CW, McConnell M, Guarner J, Shieh WJ, Malecki JM, Gerberding JL, Hughes JM, Perkins BA (2001). "Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States". Emerging Infectious Diseases. 7 (6): 933–44. doi:10.3201/eid0706.010604. PMC 2631903. PMID 11747719.
- Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P (September 2000). "Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments". Microbiology and Molecular Biology Reviews. 64 (3): 548–72. doi:10.1128/MMBR.64.3.548-572.2000. PMC 99004. PMID 10974126.
- McKenney PT, Driks A, Eichenberger P (January 2013). "The Bacillus subtilis endospore: assembly and functions of the multilayered coat". Nature Reviews. Microbiology. 11 (1): 33–44. doi:10.1038/nrmicro2921. PMID 23202530. S2CID 205498395.
- Nicholson WL, Fajardo-Cavazos P, Rebeil R, Slieman TA, Riesenman PJ, Law JF, Xue Y (August 2002). "Bacterial endospores and their significance in stress resistance". Antonie van Leeuwenhoek. 81 (1–4): 27–32. doi:10.1023/A:1020561122764. PMID 12448702. S2CID 30639022.
- Vreeland RH, Rosenzweig WD, Powers DW (October 2000). "Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal". Nature. 407 (6806): 897–900. Bibcode:2000Natur.407..897V. doi:10.1038/35038060. PMID 11057666. S2CID 9879073.
- Cano RJ, Borucki MK (May 1995). "Revival and identification of bacterial spores in 25- to 40-million-year-old Dominican amber". Science. 268 (5213): 1060–64. Bibcode:1995Sci...268.1060C. doi:10.1126/science.7538699. PMID 7538699.
- "Row over ancient bacteria". BBC News. 7 June 2001. Retrieved 26 April 2020.
- Nicholson WL, Schuerger AC, Setlow P (April 2005). "The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight". Mutation Research. 571 (1–2): 249–64. doi:10.1016/j.mrfmmm.2004.10.012. PMID 15748651.
- "Colonising the galaxy is hard. Why not send bacteria instead?". The Economist. 12 April 2018. ISSN 0013-0613. Retrieved 26 April 2020.
- Revitt-Mills SA, Vidor CJ, Watts TD, Lyras D, Rood JI, Adams V (May 2019). "Virulence Plasmids of the Pathogenic Clostridia". Microbiology Spectrum. 7 (3). doi:10.1128/microbiolspec.GPP3-0034-2018. PMID 31111816. S2CID 160013108.
- Reigadas E, van Prehn J, Falcone M, Fitzpatrick F, Vehreschild MJ, Kuijper EJ, Bouza E (July 2021). "How to: prophylactic interventions for prevention of Clostridioides difficile infection". Clinical Microbiology and Infection. 27 (12): 1777–1783. doi:10.1016/j.cmi.2021.06.037. PMID 34245901.
- Nealson KH (January 1999). "Post-Viking microbiology: new approaches, new data, new insights". Origins of Life and Evolution of the Biosphere. 29 (1): 73–93. Bibcode:1999OLEB...29...73N. doi:10.1023/A:1006515817767. PMID 11536899. S2CID 12289639.
- Xu J (June 2006). "Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances". Molecular Ecology. 15 (7): 1713–31. doi:10.1111/j.1365-294X.2006.02882.x. PMID 16689892. S2CID 16374800.
- Zillig W (December 1991). "Comparative biochemistry of Archaea and Bacteria". Current Opinion in Genetics & Development. 1 (4): 544–51. doi:10.1016/S0959-437X(05)80206-0. PMID 1822288.
- Slonczewski JL, Foster JW. Microbiology: An Evolving Science (3 ed.). WW Norton & Company. pp. 491–44.
- Hellingwerf KJ, Crielaard W, Hoff WD, Matthijs HC, Mur LR, van Rotterdam BJ (1994). "Photobiology of bacteria". Antonie van Leeuwenhoek (Submitted manuscript). 65 (4): 331–47. doi:10.1007/BF00872217. PMID 7832590. S2CID 23438926.
- Dalton H (June 2005). "The Leeuwenhoek Lecture 2000 the natural and unnatural history of methane-oxidizing bacteria". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 360 (1458): 1207–22. doi:10.1098/rstb.2005.1657. PMC 1569495. PMID 16147517.
- Imran A, Hakim S, Tariq M, Nawaz MS, Laraib I, Gulzar U, Hanif MK, Siddique MJ, Hayat M, Fraz A, Ahmad M (2021). "Diazotrophs for Lowering Nitrogen Pollution Crises: Looking Deep Into the Roots". Frontiers in Microbiology. 12: 637815. doi:10.3389/fmicb.2021.637815. PMC 8180554. PMID 34108945.
- Zehr JP, Jenkins BD, Short SM, Steward GF (July 2003). "Nitrogenase gene diversity and microbial community structure: a cross-system comparison". Environmental Microbiology. 5 (7): 539–54. doi:10.1046/j.1462-2920.2003.00451.x. PMID 12823187.
- Kosugi Y, Matsuura N, Liang Q, Yamamoto-Ikemoto R (October 2020). "Wastewater Treatment using the "Sulfate Reduction, DenitrificationAnammox and Partial Nitrification (SRDAPN)" Process". Chemosphere. 256: 127092. Bibcode:2020Chmsp.256l7092K. doi:10.1016/j.chemosphere.2020.127092. PMID 32559887. S2CID 219476361.
- Morel FM, Kraepiel AM, Amyot M (1998). "The chemical cycle and bioaccumulation of mercury". Annual Review of Ecology and Systematics. 29: 543–66. doi:10.1146/annurev.ecolsys.29.1.543. S2CID 86336987.
- Ślesak I, Kula M, Ślesak H, Miszalski Z, Strzałka K (August 2019). "How to define obligatory anaerobiosis? An evolutionary view on the antioxidant response system and the early stages of the evolution of life on Earth". Free Radical Biology & Medicine. 140: 61–73. doi:10.1016/j.freeradbiomed.2019.03.004. PMID 30862543.
- Stewart EJ, Madden R, Paul G, Taddei F (February 2005). "Aging and death in an organism that reproduces by morphologically symmetric division". PLOS Biology. 3 (2): e45. doi:10.1371/journal.pbio.0030045. PMC 546039. PMID 15685293.
- Koch AL (2002). "Control of the bacterial cell cycle by cytoplasmic growth". Critical Reviews in Microbiology. 28 (1): 61–77. doi:10.1080/1040-840291046696. PMID 12003041. S2CID 11624182.
- Pommerville 2014, p. 138.
- Pommerville 2014, p. 557.
- Wheelis 2008, p. 42.
- Thomson RB, Bertram H (December 2001). "Laboratory diagnosis of central nervous system infections". Infectious Disease Clinics of North America. 15 (4): 1047–71. doi:10.1016/S0891-5520(05)70186-0. PMID 11780267.
- Paerl HW, Fulton RS, Moisander PH, Dyble J (April 2001). "Harmful freshwater algal blooms, with an emphasis on cyanobacteria". TheScientificWorldJournal. 1: 76–113. doi:10.1100/tsw.2001.16. PMC 6083932. PMID 12805693.
- Challis GL, Hopwood DA (November 2003). "Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species". Proceedings of the National Academy of Sciences of the United States of America. 100 (Suppl 2): 14555–61. Bibcode:2003PNAS..10014555C. doi:10.1073/pnas.1934677100. PMC 304118. PMID 12970466.
- Kooijman SA, Auger P, Poggiale JC, Kooi BW (August 2003). "Quantitative steps in symbiogenesis and the evolution of homeostasis". Biological Reviews of the Cambridge Philosophical Society. 78 (3): 435–63. doi:10.1017/S1464793102006127. PMID 14558592. S2CID 41072709.
- Bertrand RL (2019). "Lag Phase is a Dynamic, Organized, Adaptive, and Evolvable Period that Prepares Bacteria for Cell Division". Journal of Bacteriology. 201 (7): e00697-18. doi:10.1128/JB.00697-18. PMC 6416914. PMID 30642990.
- Prats C, López D, Giró A, Ferrer J, Valls J (August 2006). "Individual-based modelling of bacterial cultures to study the microscopic causes of the lag phase". Journal of Theoretical Biology. 241 (4): 939–53. Bibcode:2006JThBi.241..939P. doi:10.1016/j.jtbi.2006.01.029. PMID 16524598.
- Hecker M, Völker U (2001). General stress response of Bacillus subtilis and other bacteria. Advances in Microbial Physiology. Vol. 44. pp. 35–91. doi:10.1016/S0065-2911(01)44011-2. ISBN 978-0-12-027744-5. PMID 11407115.
- Slonczewski JL, Foster JW. Microbiology: An Evolving Science (3 ed.). WW Norton & Company. p. 143.
- Leppänen M, Sundberg LR, Laanto E, de Freitas Almeida GM, Papponen P, Maasilta IJ (August 2017). "Imaging Bacterial Colonies and Phage-Bacterium Interaction at Sub-Nanometer Resolution Using Helium-Ion Microscopy". Advanced Biosystems. 1 (8): e1700070. doi:10.1002/adbi.201700070. PMID 32646179. S2CID 90960276.
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M (October 2006). "The 160-kilobase genome of the bacterial endosymbiont Carsonella". Science. 314 (5797): 267. doi:10.1126/science.1134196. PMID 17038615. S2CID 44570539.
- Pradella S, Hans A, Spröer C, Reichenbach H, Gerth K, Beyer S (December 2002). "Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56". Archives of Microbiology. 178 (6): 484–92. doi:10.1007/s00203-002-0479-2. PMID 12420170. S2CID 21023021.
- Hinnebusch J, Tilly K (December 1993). "Linear plasmids and chromosomes in bacteria". Molecular Microbiology. 10 (5): 917–22. doi:10.1111/j.1365-2958.1993.tb00963.x. PMID 7934868. S2CID 23852021.
- Lin YS, Kieser HM, Hopwood DA, Chen CW (December 1993). "The chromosomal DNA of Streptomyces lividans 66 is linear". Molecular Microbiology. 10 (5): 923–33. doi:10.1111/j.1365-2958.1993.tb00964.x. PMID 7934869. S2CID 8536066.
- Val ME, Soler-Bistué A, Bland MJ, Mazel D (December 2014). "Management of multipartite genomes: the Vibrio cholerae model". Current Opinion in Microbiology. 22: 120–26. doi:10.1016/j.mib.2014.10.003. PMID 25460805. S2CID 215743285.
- Kado CI (October 2014). "Historical events that spawned the field of plasmid biology". Microbiology Spectrum. 2 (5): 3. doi:10.1128/microbiolspec.PLAS-0019-2013. PMID 26104369.
- Belfort M, Reaban ME, Coetzee T, Dalgaard JZ (July 1995). "Prokaryotic introns and inteins: a panoply of form and function". Journal of Bacteriology. 177 (14): 3897–903. doi:10.1128/jb.177.14.3897-3903.1995. PMC 177115. PMID 7608058.
- Denamur E, Matic I (May 2006). "Evolution of mutation rates in bacteria". Molecular Microbiology. 60 (4): 820–27. doi:10.1111/j.1365-2958.2006.05150.x. PMID 16677295. S2CID 20713095.
- Wright BE (May 2004). "Stress-directed adaptive mutations and evolution". Molecular Microbiology. 52 (3): 643–50. doi:10.1111/j.1365-2958.2004.04012.x. PMID 15101972. S2CID 1071308.
- Chen I, Dubnau D (March 2004). "DNA uptake during bacterial transformation". Nature Reviews. Microbiology. 2 (3): 241–49. doi:10.1038/nrmicro844. PMID 15083159. S2CID 205499369.
- Johnsborg O, Eldholm V, Håvarstein LS (December 2007). "Natural genetic transformation: prevalence, mechanisms and function". Research in Microbiology. 158 (10): 767–78. doi:10.1016/j.resmic.2007.09.004. PMID 17997281.
- Bernstein H, Bernstein C, Michod RE (2012). "DNA repair as the primary adaptive function of sex in bacteria and eukaryotes". Chapter 1: pp. 1–49 in: DNA Repair: New Research, Sakura Kimura and Sora Shimizu (eds.). Nova Sci. Publ., Hauppauge, NY ISBN 978-1-62100-808-8.
- Brüssow H, Canchaya C, Hardt WD (September 2004). "Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion". Microbiology and Molecular Biology Reviews. 68 (3): 560–602, table of contents. doi:10.1128/MMBR.68.3.560-602.2004. PMC 515249. PMID 15353570.
- Bickle TA, Krüger DH (June 1993). "Biology of DNA restriction". Microbiological Reviews. 57 (2): 434–50. doi:10.1128/MMBR.57.2.434-450.1993. PMC 372918. PMID 8336674.
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (March 2007). "CRISPR provides acquired resistance against viruses in prokaryotes". Science. 315 (5819): 1709–12. Bibcode:2007Sci...315.1709B. doi:10.1126/science.1138140. hdl:20.500.11794/38902. PMID 17379808. S2CID 3888761.
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (August 2008). "Small CRISPR RNAs guide antiviral defense in prokaryotes". Science. 321 (5891): 960–64. Bibcode:2008Sci...321..960B. doi:10.1126/science.1159689. PMC 5898235. PMID 18703739.
- Michod RE, Bernstein H, Nedelcu AM (May 2008). "Adaptive value of sex in microbial pathogens" (PDF). Infection, Genetics and Evolution. 8 (3): 267–85. doi:10.1016/j.meegid.2008.01.002. PMID 18295550. Archived (PDF) from the original on 30 December 2016.
- Hastings PJ, Rosenberg SM, Slack A (September 2004). "Antibiotic-induced lateral transfer of antibiotic resistance". Trends in Microbiology. 12 (9): 401–14. doi:10.1016/j.tim.2004.07.003. PMID 15337159.
- Davison J (September 1999). "Genetic exchange between bacteria in the environment". Plasmid. 42 (2): 73–91. doi:10.1006/plas.1999.1421. PMID 10489325.
- Kim KW (December 2017). "Electron microscopic observations of prokaryotic surface appendages". Journal of Microbiology (Seoul, Korea). 55 (12): 919–26. doi:10.1007/s12275-017-7369-4. PMID 29214488. S2CID 89872403.
- Macnab RM (December 1999). "The bacterial flagellum: reversible rotary propellor and type III export apparatus". Journal of Bacteriology. 181 (23): 7149–53. doi:10.1128/JB.181.23.7149-7153.1999. PMC 103673. PMID 10572114.
- Wu M, Roberts JW, Kim S, Koch DL, DeLisa MP (July 2006). "Collective bacterial dynamics revealed using a three-dimensional population-scale defocused particle tracking technique". Applied and Environmental Microbiology. 72 (7): 4987–94. Bibcode:2006ApEnM..72.4987W. doi:10.1128/AEM.00158-06. PMC 1489374. PMID 16820497.
- Mattick JS (2002). "Type IV pili and twitching motility". Annual Review of Microbiology. 56: 289–314. doi:10.1146/annurev.micro.56.012302.160938. PMID 12142488.
- Merz AJ, So M, Sheetz MP (September 2000). "Pilus retraction powers bacterial twitching motility". Nature. 407 (6800): 98–102. Bibcode:2000Natur.407...98M. doi:10.1038/35024105. PMID 10993081. S2CID 4425775.
- Lux R, Shi W (July 2004). "Chemotaxis-guided movements in bacteria". Critical Reviews in Oral Biology and Medicine. 15 (4): 207–20. doi:10.1177/154411130401500404. PMID 15284186.
- Schweinitzer T, Josenhans C (July 2010). "Bacterial energy taxis: a global strategy?". Archives of Microbiology. 192 (7): 507–20. doi:10.1007/s00203-010-0575-7. PMC 2886117. PMID 20411245.
- Frankel RB, Bazylinski DA, Johnson MS, Taylor BL (August 1997). "Magneto-aerotaxis in marine coccoid bacteria". Biophysical Journal. 73 (2): 994–1000. Bibcode:1997BpJ....73..994F. doi:10.1016/S0006-3495(97)78132-3. PMC 1180996. PMID 9251816.
- Nan B, Zusman DR (2011). "Uncovering the mystery of gliding motility in the myxobacteria". Annual Review of Genetics. 45: 21–39. doi:10.1146/annurev-genet-110410-132547. PMC 3397683. PMID 21910630.
- Goldberg MB (December 2001). "Actin-based motility of intracellular microbial pathogens". Microbiology and Molecular Biology Reviews. 65 (4): 595–626, table of contents. doi:10.1128/MMBR.65.4.595-626.2001. PMC 99042. PMID 11729265.
- Calcagnile M, Tredici SM, Talà A, Alifano P (December 2019). "Bacterial Semiochemicals and Transkingdom Interactions with Insects and Plants". Insects. 10 (12): 441. doi:10.3390/insects10120441. PMC 6955855. PMID 31817999.
- Shapiro JA (1998). "Thinking about bacterial populations as multicellular organisms" (PDF). Annual Review of Microbiology. 52: 81–104. doi:10.1146/annurev.micro.52.1.81. PMID 9891794. Archived from the original (PDF) on 17 July 2011.
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995). "Microbial biofilms". Annual Review of Microbiology. 49: 711–45. doi:10.1146/annurev.mi.49.100195.003431. PMID 8561477.
- Miller DP, Lamont RJ (2019). "Signaling systems in oral bacteria". Advances in Experimental Medicine and Biology. 1197: 27–43. doi:10.1007/978-3-030-28524-1_3. ISBN 978-3-030-28523-4. PMID 31732932. S2CID 208063186.
- Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR (May 2018). "Bacterial Quorum sensing and microbial community interactions". mBio. 9 (3). doi:10.1128/mBio.02331-17. PMC 5964356. PMID 29789364.
- Miller MB, Bassler BL (2001). "Quorum sensing in bacteria". Annual Review of Microbiology. 55: 165–99. doi:10.1146/annurev.micro.55.1.165. PMID 11544353. S2CID 1099089.
- Zhu Q, Mai U, Pfeiffer W, Janssen S, Asnicar F, Sanders JG, et al. (December 2019). "Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea". Nature Communications. 10 (1): 5477. Bibcode:2019NatCo..10.5477Z. doi:10.1038/s41467-019-13443-4. PMC 6889312. PMID 31792218.
- Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbø CL, Case RJ, Doolittle WF (2003). "Lateral gene transfer and the origins of prokaryotic groups". Annual Review of Genetics. 37: 283–328. doi:10.1146/annurev.genet.37.050503.084247. PMID 14616063.
- Olsen GJ, Woese CR, Overbeek R (January 1994). "The winds of (evolutionary) change: breathing new life into microbiology". Journal of Bacteriology. 176 (1): 1–6. doi:10.2172/205047. PMC 205007. PMID 8282683.
- "IJSEM Home". Ijs.sgmjournals.org. 28 October 2011. Archived from the original on 19 October 2011. Retrieved 4 November 2011.
- "Bergey's Manual Trust". Bergeys.org. Archived from the original on 7 November 2011. Retrieved 4 November 2011.
- Hedlund BP, Dodsworth JA, Staley JT (June 2015). "The changing landscape of microbial biodiversity exploration and its implications for systematics". Systematic and Applied Microbiology. 38 (4): 231–36. doi:10.1016/j.syapm.2015.03.003. PMID 25921438.
- "Schizomycetes." Merriam-Webster.com Medical Dictionary, Merriam-Webster, https://www.merriam-webster.com/medical/Schizomycetes. Accessed 3 August 2021.
- Brown MM, Horswill AR (November 2020). "Staphylococcus epidermidis-Skin friend or foe?". PLOS Pathogens. 16 (11): e1009026. doi:10.1371/journal.ppat.1009026. PMC 7660545. PMID 33180890.
- Gupta RS (2000). "The natural evolutionary relationships among prokaryotes". Critical Reviews in Microbiology. 26 (2): 111–31. CiteSeerX 10.1.1.496.1356. doi:10.1080/10408410091154219. PMID 10890353. S2CID 30541897.
- Rappé MS, Giovannoni SJ (2003). "The uncultured microbial majority". Annual Review of Microbiology. 57: 369–94. doi:10.1146/annurev.micro.57.030502.090759. PMID 14527284. S2CID 10781051.
- Doolittle RF (June 2005). "Evolutionary aspects of whole-genome biology". Current Opinion in Structural Biology. 15 (3): 248–53. doi:10.1016/j.sbi.2005.04.001. PMID 15963888.
- Cavalier-Smith T (January 2002). "The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification". International Journal of Systematic and Evolutionary Microbiology. 52 (Pt 1): 7–76. doi:10.1099/00207713-52-1-7. PMID 11837318.
- Pommerville 2014, p. 15−31.
- Krasner 2014, p. 77.
- Woods GL, Walker DH (July 1996). "Detection of infection or infectious agents by use of cytologic and histologic stains". Clinical Microbiology Reviews. 9 (3): 382–404. doi:10.1128/CMR.9.3.382. PMC 172900. PMID 8809467.
- Tang S, Orsi RH, Luo H, Ge C, Zhang G, Baker RC, Stevenson A, Wiedmann M (2019). "Assessment and Comparison of Molecular Subtyping and Characterization Methods for Salmonella". Frontiers in Microbiology. 10: 1591. doi:10.3389/fmicb.2019.01591. PMC 6639432. PMID 31354679.
- Krasner 2014, p. 87–89.
- Weinstein MP (March 1994). "Clinical importance of blood cultures". Clinics in Laboratory Medicine. 14 (1): 9–16. doi:10.1016/S0272-2712(18)30390-1. PMID 8181237.
- Riley LW (November 2018). "Laboratory Methods in Molecular Epidemiology: Bacterial Infections". Microbiology Spectrum. 6 (6). doi:10.1128/microbiolspec.AME-0004-2018. PMID 30387415. S2CID 54234977.
- Lenkowski M, Nijakowski K, Kaczmarek M, Surdacka A (March 2021). "The Loop-Mediated Isothermal Amplification Technique in Periodontal Diagnostics: A Systematic Review". Journal of Clinical Medicine. 10 (6): 1189. doi:10.3390/jcm10061189. PMC 8000232. PMID 33809163.
- Alizadeh M, Yousefi L, Pakdel F, Ghotaslou R, Rezaee MA, Khodadadi E, Oskouei MA, Soroush Barhaghi MH, Kafil HS (2021). "MALDI-TOF Mass Spectroscopy Applications in Clinical Microbiology". Advances in Pharmacological and Pharmaceutical Sciences. 2021: 9928238. doi:10.1155/2021/9928238. PMC 8121603. PMID 34041492.
- Dudek NK, Sun CL, Burstein D (2017). "Novel Microbial Diversity and Functional Potential in the Marine Mammal Oral Microbiome" (PDF). Current Biology. 27 (24): 3752–62. doi:10.1016/j.cub.2017.10.040. PMID 29153320. S2CID 43864355.
- Louie M, Louie L, Simor AE (August 2000). "The role of DNA amplification technology in the diagnosis of infectious diseases". CMAJ. 163 (3): 301–09. doi:10.1016/s1381-1169(00)00220-x. PMC 80298. PMID 10951731. Archived from the original on 14 June 2006.
- Oliver JD (February 2005). "The viable but nonculturable state in bacteria". Journal of Microbiology. 43 Spec No: 93–100. PMID 15765062. Archived from the original on 28 September 2007.
- Euzéby JP (8 December 2011). "Number of published names". List of Prokaryotic names with Standing in Nomenclature. Archived from the original on 19 January 2012. Retrieved 10 December 2011.
- Curtis TP, Sloan WT, Scannell JW (August 2002). "Estimating prokaryotic diversity and its limits". Proceedings of the National Academy of Sciences of the United States of America. 99 (16): 10494–99. Bibcode:2002PNAS...9910494C. doi:10.1073/pnas.142680199. PMC 124953. PMID 12097644.
- Schloss PD, Handelsman J (December 2004). "Status of the microbial census". Microbiology and Molecular Biology Reviews. 68 (4): 686–91. doi:10.1128/MMBR.68.4.686-691.2004. PMC 539005. PMID 15590780.
- Oren A, Garrity GM (2021). "Valid publication of the names of forty-two phyla of prokaryotes". Int J Syst Evol Microbiol. 71 (10): 5056. doi:10.1099/ijsem.0.005056. PMID 34694987. S2CID 239887308.
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. (2013). "Insights into the phylogeny and coding potential of microbial dark matter". Nature. 499 (7459): 431–437. Bibcode:2013Natur.499..431R. doi:10.1038/nature12352. PMID 23851394. S2CID 4394530.
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. (2015). "Unusual biology across a group comprising more than 15% of domain Bacteria". Nature. 523 (7559): 208–211. Bibcode:2015Natur.523..208B. doi:10.1038/nature14486. OSTI 1512215. PMID 26083755. S2CID 4397558.
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. (2016). "A new view of the tree of life". Nat Microbiol. 1 (5): 16048. doi:10.1038/nmicrobiol.2016.48. PMID 27572647. S2CID 3833474.
- Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, Brodie EL, Williams KH, Hubbard SS, Banfield JF. (2016). "Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system". Nat Commun. 7: 13219. Bibcode:2016NatCo...713219A. doi:10.1038/ncomms13219. PMC 5079060. PMID 27774985.
- Fisher B, Harvey RP, Champe PC (2007). "Chapter 33". Lippincott's Illustrated Reviews: Microbiology (Lippincott's Illustrated Reviews Series). Hagerstwon, MD: Lippincott Williams & Wilkins. pp. 367–92. ISBN 978-0-7817-8215-9.
- Mushegian AA, Ebert D (January 2016). "Rethinking "mutualism" in diverse host-symbiont communities". BioEssays. 38 (1): 100–8. doi:10.1002/bies.201500074. PMID 26568407. S2CID 31661712.
- Harper, Douglas. "commensalism". Online Etymology Dictionary.
- Sears CL (October 2005). "A dynamic partnership: celebrating our gut flora". Anaerobe. 11 (5): 247–51. doi:10.1016/j.anaerobe.2005.05.001. PMID 16701579.
- Khan R, Petersen FC, Shekhar S (2019). "Commensal Bacteria: An Emerging Player in Defense Against Respiratory Pathogens". Frontiers in Immunology. 10: 1203. doi:10.3389/fimmu.2019.01203. PMC 6554327. PMID 31214175.
- Roscoe DL, Chow AW (March 1988). "Normal flora and mucosal immunity of the head and neck". Infectious Disease Clinics of North America. 2 (1): 1–19. doi:10.1016/S0891-5520(20)30163-X. PMID 3074102.
- Shiao SL, Kershaw KM, Limon JJ, You S, Yoon J, Ko EY, Guarnerio J, Potdar AA, McGovern DP, Bose S, Dar TB, Noe P, Lee J, Kubota Y, Maymi VI, Davis MJ, Henson RM, Choi RY, Yang W, Tang J, Gargus M, Prince AD, Zumsteg ZS, Underhill DM (July 2021). "Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy". Cancer Cell. 39 (9): 1202–1213.e6. doi:10.1016/j.ccell.2021.07.002. PMC 8830498. PMID 34329585.
- Lyapichev KA, Ivashkevich Y, Chernov Y, Chinenov D, Shpot E, Bessonov AA, Dabaja BS, Konoplev S (2021). "MALT Lymphoma of the Urinary Bladder Shows a Dramatic Female Predominance, Uneven Geographic Distribution, and Possible Infectious Etiology". Research and Reports in Urology. 13: 49–62. doi:10.2147/RRU.S283366. PMC 7873029. PMID 33575225.
- Eleyan L, Khan AA, Musollari G, Chandiramani AS, Shaikh S, Salha A, Tarmahomed A, Harky A (April 2021). "Infective endocarditis in paediatric population". European Journal of Pediatrics. 180 (10): 3089–3100. doi:10.1007/s00431-021-04062-7. PMID 33852085. S2CID 233225250.
- Martin MO (September 2002). "Predatory prokaryotes: an emerging research opportunity". Journal of Molecular Microbiology and Biotechnology. 4 (5): 467–77. PMID 12432957.
- Velicer GJ, Stredwick KL (August 2002). "Experimental social evolution with Myxococcus xanthus". Antonie van Leeuwenhoek. 81 (1–4): 155–64. doi:10.1023/A:1020546130033. PMID 12448714. S2CID 20018104.
- Bauer A, Forchhammer K (May 2021). "Bacterial Predation on Cyanobacteria". Microbial Physiology. 31 (2): 99–108. doi:10.1159/000516427. ISSN 2673-1665. PMID 34010833.
- Velicer GJ, Mendes-Soares H (January 2009). "Bacterial predators". Current Biology. 19 (2): R55–56. doi:10.1016/j.cub.2008.10.043. PMID 19174136. S2CID 5432036.
- Stams AJ, de Bok FA, Plugge CM, van Eekert MH, Dolfing J, Schraa G (March 2006). "Exocellular electron transfer in anaerobic microbial communities". Environmental Microbiology. 8 (3): 371–82. doi:10.1111/j.1462-2920.2006.00989.x. PMID 16478444.
- Zinser ER (August 2018). "Cross-protection from hydrogen peroxide by helper microbes: the impacts on the cyanobacterium Prochlorococcus and other beneficiaries in marine communities". Environmental Microbiology Reports. 10 (4): 399–411. doi:10.1111/1758-2229.12625. PMID 29411546.
- Barea JM, Pozo MJ, Azcón R, Azcón-Aguilar C (July 2005). "Microbial co-operation in the rhizosphere". Journal of Experimental Botany. 56 (417): 1761–78. doi:10.1093/jxb/eri197. PMID 15911555.
- O'Hara AM, Shanahan F (July 2006). "The gut flora as a forgotten organ". EMBO Reports. 7 (7): 688–93. doi:10.1038/sj.embor.7400731. PMC 1500832. PMID 16819463.
- Zoetendal EG, Vaughan EE, de Vos WM (March 2006). "A microbial world within us". Molecular Microbiology. 59 (6): 1639–50. doi:10.1111/j.1365-2958.2006.05056.x. PMID 16553872. S2CID 37602619.
- Gorbach SL (February 1990). "Lactic acid bacteria and human health". Annals of Medicine. 22 (1): 37–41. doi:10.3109/07853899009147239. PMID 2109988.
- Salminen SJ, Gueimonde M, Isolauri E (May 2005). "Probiotics that modify disease risk". The Journal of Nutrition. 135 (5): 1294–98. doi:10.1093/jn/135.5.1294. PMID 15867327.
- Watanabe F, Bito T (January 2018). "Vitamin B12 sources and microbial interaction". Experimental Biology and Medicine. 243 (2): 148–58. doi:10.1177/1535370217746612. PMC 5788147. PMID 29216732.
- Pommerville 2014, pp. 16–21.
- Clark 2010, p. 215.
- Wheelis 2008, p. 44.
- Clark 2010, pp. 30, 195, 233, 236.
- Miller AK, Williams SM (July 2021). "Helicobacter pylori infection causes both protective and deleterious effects in human health and disease". Genes and Immunity. 22 (4): 218–226. doi:10.1038/s41435-021-00146-4. PMC 8390445. PMID 34244666.
- Schwarz S, Enne VI, van Duijkeren E (October 2016). "40 years of veterinary papers in JAC – what have we learnt?". The Journal of Antimicrobial Chemotherapy. 71 (10): 2681–90. doi:10.1093/jac/dkw363. PMID 27660260.
- Fish DN (February 2002). "Optimal antimicrobial therapy for sepsis". American Journal of Health-System Pharmacy. 59 (Suppl 1): S13–19. doi:10.1093/ajhp/59.suppl_1.S13. PMID 11885408.
- Belland RJ, Ouellette SP, Gieffers J, Byrne GI (February 2004). "Chlamydia pneumoniae and atherosclerosis". Cellular Microbiology. 6 (2): 117–27. doi:10.1046/j.1462-5822.2003.00352.x. PMID 14706098. S2CID 45218449.
- Heise ER (February 1982). "Diseases associated with immunosuppression". Environmental Health Perspectives. 43: 9–19. doi:10.2307/3429162. JSTOR 3429162. PMC 1568899. PMID 7037390.
- Saiman L (2004). "Microbiology of early CF lung disease". Paediatric Respiratory Reviews. 5 (Suppl A): S367–69. doi:10.1016/S1526-0542(04)90065-6. PMID 14980298.
- Pommerville 2014, p. 118.
- Pommerville 2014, pp. 646–47.
- Krasner 2014, pp. 165, 369.
- Yonath A, Bashan A (2004). "Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics". Annual Review of Microbiology. 58: 233–51. doi:10.1146/annurev.micro.58.030603.123822. PMID 15487937.
- Khachatourians GG (November 1998). "Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria". CMAJ. 159 (9): 1129–36. PMC 1229782. PMID 9835883.
- Kuo J (October 2017). "Disinfection Processes". Water Environment Research. 89 (10): 1206–44. doi:10.2175/106143017X15023776270278. PMID 28954657.
- Johnson ME, Lucey JA (April 2006). "Major technological advances and trends in cheese". Journal of Dairy Science. 89 (4): 1174–78. doi:10.3168/jds.S0022-0302(06)72186-5. PMID 16537950.
- Krasner 2014, pp. 25–26.
- Cohen Y (December 2002). "Bioremediation of oil by marine microbial mats". International Microbiology. 5 (4): 189–93. doi:10.1007/s10123-002-0089-5. PMID 12497184. S2CID 26039323.
- Neves LC, Miyamura TT, Moraes DA, Penna TC, Converti A (2006). "Biofiltration methods for the removal of phenolic residues". Applied Biochemistry and Biotechnology. 129–132 (1–3): 130–52. doi:10.1385/ABAB:129:1:130. PMID 16915636. S2CID 189905816.
- Liese A, Filho MV (December 1999). "Production of fine chemicals using biocatalysis". Current Opinion in Biotechnology. 10 (6): 595–603. doi:10.1016/S0958-1669(99)00040-3. PMID 10600695.
- Aronson AI, Shai Y (February 2001). "Why Bacillus thuringiensis insecticidal toxins are so effective: unique features of their mode of action". FEMS Microbiology Letters. 195 (1): 1–8. doi:10.1111/j.1574-6968.2001.tb10489.x. PMID 11166987.
- Bozsik A (July 2006). "Susceptibility of adult Coccinella septempunctata (Coleoptera: Coccinellidae) to insecticides with different modes of action". Pest Management Science. 62 (7): 651–54. doi:10.1002/ps.1221. PMID 16649191.
- Chattopadhyay A, Bhatnagar NB, Bhatnagar R (2004). "Bacterial insecticidal toxins". Critical Reviews in Microbiology. 30 (1): 33–54. doi:10.1080/10408410490270712. PMID 15116762. S2CID 1580984.
- Serres MH, Gopal S, Nahum LA, Liang P, Gaasterland T, Riley M (2001). "A functional update of the Escherichia coli K-12 genome". Genome Biology. 2 (9): RESEARCH0035. doi:10.1186/gb-2001-2-9-research0035. PMC 56896. PMID 11574054.
- Almaas E, Kovács B, Vicsek T, Oltvai ZN, Barabási AL (February 2004). "Global organization of metabolic fluxes in the bacterium Escherichia coli". Nature. 427 (6977): 839–43. arXiv:q-bio/0403001. Bibcode:2004Natur.427..839A. doi:10.1038/nature02289. PMID 14985762. S2CID 715721.
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003). "An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR)". Genome Biology. 4 (9): R54. doi:10.1186/gb-2003-4-9-r54. PMC 193654. PMID 12952533.
- Walsh G (April 2005). "Therapeutic insulins and their large-scale manufacture". Applied Microbiology and Biotechnology. 67 (2): 151–59. doi:10.1007/s00253-004-1809-x. PMID 15580495. S2CID 5986035.
- Graumann K, Premstaller A (February 2006). "Manufacturing of recombinant therapeutic proteins in microbial systems". Biotechnology Journal. 1 (2): 164–86. doi:10.1002/biot.200500051. PMID 16892246. S2CID 24702839.
- Rabsch W, Helm RA, Eisenstark A (February 2004). "Diversity of phage types among archived cultures of the Demerec collection of Salmonella enterica serovar Typhimurium strains". Applied and Environmental Microbiology. 70 (2): 664–69. Bibcode:2004ApEnM..70..664R. doi:10.1128/aem.70.2.664-669.2004. PMC 348941. PMID 14766539.
- Wheelis 2008.
- Asimov I (1982). Asimov's Biographical Encyclopedia of Science and Technology (2nd ed.). Garden City, NY: Doubleday and Company. p. 143].
- Pommerville 2014, p. 7.
- Ehrenberg CG (1828). Symbolae Physioe. Animalia evertebrata. Berlin: Decas prima.
- Breed RS, Conn HJ (May 1936). "The Status of the Generic Term Bacterium Ehrenberg 1828". Journal of Bacteriology. 31 (5): 517–18. doi:10.1128/jb.31.5.517-518.1936. PMC 543738. PMID 16559906.
- Ehrenberg CG (1835). Dritter Beitrag zur Erkenntniss grosser Organisation in der Richtung des kleinsten Raumes [Third contribution to the knowledge of great organization in the direction of the smallest space] (in German). Berlin: Physikalische Abhandlungen der Koeniglichen Akademie der Wissenschaften. pp. 143–336.
- "Pasteur's Papers on the Germ Theory". LSU Law Center's Medical and Public Health Law Site, Historic Public Health Articles. Archived from the original on 18 December 2006. Retrieved 23 November 2006.
- 'Wash your hands' was once controversial medical advice, National Geographic.
- "The Nobel Prize in Physiology or Medicine 1905". Nobelprize.org. Archived from the original on 10 December 2006. Retrieved 22 November 2006.
- O'Brien SJ, Goedert JJ (October 1996). "HIV causes AIDS: Koch's postulates fulfilled". Current Opinion in Immunology. 8 (5): 613–18. doi:10.1016/S0952-7915(96)80075-6. PMID 8902385.
- Chung KT. "Ferdinand Julius Cohn (1828–1898): Pioneer of Bacteriology" (PDF). Department of Microbiology and Molecular Cell Sciences, The University of Memphis. Archived (PDF) from the original on 27 July 2011.
- Drews, Gerhart (1999). "Ferdinand Cohn, a founder of modern microbiology" (PDF). ASM News. 65 (8): 547–52. Archived from the original (PDF) on 13 July 2017.
- Thurston AJ (December 2000). "Of blood, inflammation and gunshot wounds: the history of the control of sepsis". The Australian and New Zealand Journal of Surgery. 70 (12): 855–61. doi:10.1046/j.1440-1622.2000.01983.x. PMID 11167573.
- Schwartz RS (March 2004). "Paul Ehrlich's magic bullets". The New England Journal of Medicine. 350 (11): 1079–80. doi:10.1056/NEJMp048021. PMID 15014180.
- "Biography of Paul Ehrlich". Nobelprize.org. Archived from the original on 28 November 2006. Retrieved 26 November 2006.
- Woese CR, Fox GE (November 1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proceedings of the National Academy of Sciences of the United States of America. 74 (11): 5088–90. Bibcode:1977PNAS...74.5088W. doi:10.1073/pnas.74.11.5088. PMC 432104. PMID 270744.
Bibliography
- Clark D (2010). Germs, Genes, & Civilization : how epidemics shaped who we are today. Upper Saddle River, N.J: FT Press. ISBN 978-0-13-701996-0. OCLC 473120711.
- Crawford D (2007). Deadly Companions : how microbes shaped our history. Oxford New York: Oxford University Press. ISBN 978-0-19-956144-5. OCLC 183198723.
- Hall B (2008). Strickberger's Evolution : the integration of genes, organisms and populations. Sudbury, Mass: Jones and Bartlett. ISBN 978-0-7637-0066-9. OCLC 85814089.
- Krasner R (2014). The Microbial Challenge : a public health perspective. Burlington, Mass: Jones & Bartlett Learning. ISBN 978-1-4496-7375-8. OCLC 794228026.
- Pommerville JC (2014). Fundamentals of Microbiology (10th ed.). Boston: Jones and Bartlett. ISBN 978-1-284-03968-9.
- Wheelis M (2008). Principles of modern microbiology. Sudbury, Mass: Jones and Bartlett Publishers. ISBN 978-0-7637-1075-0. OCLC 67392796.