Biology
Biology is the scientific study of life.[1][2][3] It is a natural science with a broad scope but has several unifying themes that tie it together as a single, coherent field.[1][2][3] For instance, all organisms are made up of cells that process hereditary information encoded in genes, which can be transmitted to future generations. Another major theme is evolution, which explains the unity and diversity of life.[1][2][3] Energy processing is also important to life as it allows organisms to move, grow, and reproduce.[1][2][3] Finally, all organisms are able to regulate their own internal environments.[1][2][3][4][5]
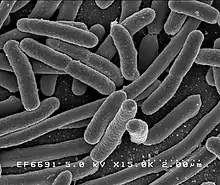
- top: E. coli bacteria and gazelle
- bottom: Goliath beetle and tree fern
| Part of a series on |
| Biology |
|---|
|
Biologists are able to study life at multiple levels of organization,[1] from the molecular biology of a cell to the anatomy and physiology of plants and animals, and evolution of populations.[1][6] Hence, there are multiple subdisciplines within biology, each defined by the nature of their research questions and the tools that they use.[7][8][9] Like other scientists, biologists use the scientific method to make observations, pose questions, generate hypotheses, perform experiments, and form conclusions about the world around them.[1]
Life on Earth, which emerged more than 3.7 billion years ago,[10] is immensely diverse. Biologists have sought to study and classify the various forms of life, from prokaryotic organisms such as archaea and bacteria to eukaryotic organisms such as protists, fungi, plants, and animals. These various organisms contribute to the biodiversity of an ecosystem, where they play specialized roles in the cycling of nutrients and energy through their biophysical environment.
History

The earliest of roots of science, which included medicine, can be traced to ancient Egypt and Mesopotamia in around 3000 to 1200 BCE.[11][12] Their contributions later entered and shaped Greek natural philosophy of classical antiquity.[11][12][13][14] Ancient Greek philosophers such as Aristotle (384–322 BCE) contributed extensively to the development of biological knowledge. His works such as History of Animals were especially important because they revealed his naturalist leanings, and later more empirical works that focused on biological causation and the diversity of life. Aristotle's successor at the Lyceum, Theophrastus, wrote a series of books on botany that survived as the most important contribution of antiquity to the plant sciences, even into the Middle Ages.[15]
Scholars of the medieval Islamic world who wrote on biology included al-Jahiz (781–869), Al-Dīnawarī (828–896), who wrote on botany,[16] and Rhazes (865–925) who wrote on anatomy and physiology. Medicine was especially well studied by Islamic scholars working in Greek philosopher traditions, while natural history drew heavily on Aristotelian thought, especially in upholding a fixed hierarchy of life.
Biology began to quickly develop and grow with Anton van Leeuwenhoek's dramatic improvement of the microscope. It was then that scholars discovered spermatozoa, bacteria, infusoria and the diversity of microscopic life. Investigations by Jan Swammerdam led to new interest in entomology and helped to develop the basic techniques of microscopic dissection and staining.[17]
Advances in microscopy also had a profound impact on biological thinking. In the early 19th century, a number of biologists pointed to the central importance of the cell. Then, in 1838, Schleiden and Schwann began promoting the now universal ideas that (1) the basic unit of organisms is the cell and (2) that individual cells have all the characteristics of life, although they opposed the idea that (3) all cells come from the division of other cells. However, Robert Remak and Rudolf Virchow were able to reify the third tenet, and by the 1860s most biologists accepted all three tenets which consolidated into cell theory.[18][19]
Meanwhile, taxonomy and classification became the focus of natural historians. Carl Linnaeus published a basic taxonomy for the natural world in 1735 (variations of which have been in use ever since), and in the 1750s introduced scientific names for all his species.[20] Georges-Louis Leclerc, Comte de Buffon, treated species as artificial categories and living forms as malleable—even suggesting the possibility of common descent. Although he was opposed to evolution, Buffon is a key figure in the history of evolutionary thought; his work influenced the evolutionary theories of both Lamarck and Darwin.[21]

Serious evolutionary thinking originated with the works of Jean-Baptiste Lamarck, who was the first to present a coherent theory of evolution.[23] He posited that evolution was the result of environmental stress on properties of animals, meaning that the more frequently and rigorously an organ was used, the more complex and efficient it would become, thus adapting the animal to its environment. Lamarck believed that these acquired traits could then be passed on to the animal's offspring, who would further develop and perfect them.[24] However, it was the British naturalist Charles Darwin, combining the biogeographical approach of Humboldt, the uniformitarian geology of Lyell, Malthus's writings on population growth, and his own morphological expertise and extensive natural observations, who forged a more successful evolutionary theory based on natural selection; similar reasoning and evidence led Alfred Russel Wallace to independently reach the same conclusions.[25][26] Darwin's theory of evolution by natural selection quickly spread through the scientific community and soon became a central axiom of the rapidly developing science of biology.
The basis for modern genetics began with the work of Gregor Mendel, who presented his paper, "Versuche über Pflanzenhybriden" ("Experiments on Plant Hybridization"), in 1865,[27] which outlined the principles of biological inheritance, serving as the basis for modern genetics.[28] However, the significance of his work was not realized until the early 20th century when evolution became a unified theory as the modern synthesis reconciled Darwinian evolution with classical genetics.[29] In the 1940s and early 1950s, a series of experiments by Alfred Hershey and Martha Chase pointed to DNA as the component of chromosomes that held the trait-carrying units that had become known as genes. A focus on new kinds of model organisms such as viruses and bacteria, along with the discovery of the double-helical structure of DNA by James Watson and Francis Crick in 1953, marked the transition to the era of molecular genetics. From the 1950s onwards, biology has been vastly extended in the molecular domain. The genetic code was cracked by Har Gobind Khorana, Robert W. Holley and Marshall Warren Nirenberg after DNA was understood to contain codons. Finally, the Human Genome Project was launched in 1990 with the goal of mapping the general human genome. This project was essentially completed in 2003,[30] with further analysis still being published. The Human Genome Project was the first step in a globalized effort to incorporate accumulated knowledge of biology into a functional, molecular definition of the human body and the bodies of other organisms.
Chemical basis
Atoms and molecules
All organisms are made up of chemical elements;[31] oxygen, carbon, hydrogen, and nitrogen account for 96% of all organisms, with calcium, phosphorus, sulfur, sodium, chlorine, and magnesium constituting essentially all the remainder. Different elements can combine to form compounds such as water, which is fundamental to life.[31] Biochemistry is the study of chemical processes within and relating to living organisms. Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including molecular synthesis, modification, mechanisms, and interactions.
Water

Life arose from the Earth's first ocean, which was formed approximately 3.8 billion years ago.[32] Since then, water continues to be the most abundant molecule in every organism. Water is important to life because it is an effective solvent, capable of dissolving solutes such as sodium and chloride ions or other small molecules to form an aqueous solution. Once dissolved in water, these solutes are more likely to come in contact with one another and therefore take part in chemical reactions that sustain life.[32]
In terms of its molecular structure, water is a small polar molecule with a bent shape formed by the polar covalent bonds of two hydrogen (H) atoms to one oxygen (O) atom (H2O).[32] Because the O–H bonds are polar, the oxygen atom has a slight negative charge and the two hydrogen atoms have a slight positive charge.[32] This polar property of water allows it to attract other water molecules via hydrogen bonds, which makes water cohesive.[32] Surface tension results from the cohesive force due to the attraction between molecules at the surface of the liquid.[32] Water is also adhesive as it is able to adhere to the surface of any polar or charged non-water molecules.[32]
Water is denser as a liquid than it is as a solid (or ice).[32] This unique property of water allows ice to float above liquid water such as ponds, lakes, and oceans, thereby insulating the liquid below from the cold air above.[32] The lower density of ice compared to liquid water is due to the lower number of water molecules that form the crystal lattice structure of ice, which leaves a large amount of space between water molecules.[32] In contrast, there is no crystal lattice structure in liquid water, which allows more water molecules to occupy the same amount of volume.[32]
Water also has the capacity to absorb energy, giving it a higher specific heat capacity than other solvents such as ethanol.[32] Thus, a large amount of energy is needed to break the hydrogen bonds between water molecules to convert liquid water into water vapor.[32]
As a molecule, water is not completely stable as each water molecule continuously dissociates into hydrogen and hydroxyl ions before reforming into a water molecule again.[32] In pure water, the number of hydrogen ions balances (or equals) the number of hydroxyl ions, resulting in a pH that is neutral.
Organic compounds

Organic compounds are molecules that contain carbon bonded to another element such as hydrogen.[32] With the exception of water, nearly all the molecules that make up each organism contain carbon.[32][33] Carbon can form covalent bonds with up to four other atoms, enabling it to form diverse, large, and complex molecules.[32][33] For example, a single carbon atom can form four single covalent bonds such as in methane, two double covalent bonds such as in carbon dioxide (CO2), or a triple covalent bond such as in carbon monoxide (CO). Moreover, carbon can form very long chains of interconnecting carbon–carbon bonds such as octane or ring-like structures such as glucose.
The simplest form of an organic molecule is the hydrocarbon, which is a large family of organic compounds that are composed of hydrogen atoms bonded to a chain of carbon atoms. A hydrocarbon backbone can be substituted by other elements such as oxygen (O), hydrogen (H), phosphorus (P), and sulfur (S), which can change the chemical behavior of that compound.[32] Groups of atoms that contain these elements (O-, H-, P-, and S-) and are bonded to a central carbon atom or skeleton are called functional groups.[32] There are six prominent functional groups that can be found in organisms: amino group, carboxyl group, carbonyl group, hydroxyl group, phosphate group, and sulfhydryl group.[32]
In 1953, the Miller-Urey experiment showed that organic compounds could be synthesized abiotically within a closed system mimicking the conditions of early Earth, thus suggesting that complex organic molecules could have arisen spontaneously in early Earth (see abiogenesis).[34][32]
Macromolecules

Macromolecules are large molecules made up of smaller molecular subunits that are joined.[35] Small molecules such as sugars, amino acids, and nucleotides can act as single repeating units called monomers to form chain-like molecules called polymers via a chemical process called condensation.[36] For example, amino acids can form polypeptides whereas nucleotides can form strands of nucleic acid. Polymers make up three of the four macromolecules (polysaccharides, lipids, proteins, and nucleic acids) that are found in all organisms. Each of these macromolecules plays a specialized role within any given cell.
Carbohydrates (or sugar) are molecules with the molecular formula (CH2O)n, with n being the number of carbon-hydrate groups.[37] They include monosaccharides (monomer), oligosaccharides (small polymers), and polysaccharides (large polymers). Monosaccharides can be linked together by glycosidic linkages, a type of covalent bond.[37] When two monosaccharides such as glucose and fructose are linked together, they can form a disaccharide such as sucrose.[37] When many monosaccharides are linked together, they can form an oligosaccharide or a polysaccharide, depending on the number of monosaccharides. Polysaccharides can vary in function. Monosaccharides such as glucose can be a source of energy and some polysaccharides can serve as storage material that can be hydrolyzed to provide cells with sugar.
Lipids are the only class of macromolecules that are not made up of polymers. The most biologically important lipids are steroids, phospholipids, and fats.[36] These lipids are organic compounds that are largely nonpolar and hydrophobic.[38] Steroids are organic compounds that consist of four fused rings.[38] Phospholipids consist of glycerol that is linked to a phosphate group and two hydrocarbon chains (or fatty acids).[38] The glycerol and phosphate group together constitute the polar and hydrophilic (or head) region of the molecule whereas the fatty acids make up the nonpolar and hydrophobic (or tail) region.[38] Thus, when in water, phospholipids tend to form a phospholipid bilayer whereby the hydrophobic heads face outwards to interact with water molecules. Conversely, the hydrophobic tails face inwards towards other hydrophobic tails to avoid contact with water.[38]

Proteins are the most diverse of the macromolecules, which include enzymes, transport proteins, large signaling molecules, antibodies, and structural proteins. The basic unit (or monomer) of a protein is an amino acid, which has a central carbon atom that is covalently bonded to a hydrogen atom, an amino group, a carboxyl group, and a side chain (or R-group, "R" for residue).[35] There are twenty amino acids that make up the building blocks of proteins, with each amino acid having its own unique side chain.[35] The polarity and charge of the side chains affect the solubility of amino acids. An amino acid with a side chain that is polar and electrically charged is soluble as it is hydrophilic whereas an amino acid with a side chain that lacks a charged or an electronegative atom is hydrophobic and therefore tends to coalesce rather than dissolve in water.[35] Proteins have four distinct levels of organization (primary, secondary, tertiary, and quartenary). The primary structure consists of a unique sequence of amino acids that are covalently linked together by peptide bonds.[35] The side chains of the individual amino acids can then interact with each other, giving rise to the secondary structure of a protein.[35] The two common types of secondary structures are alpha helices and beta sheets.[35] The folding of alpha helices and beta sheets gives a protein its three-dimensional or tertiary structure. Finally, multiple tertiary structures can combine to form the quaternary structure of a protein.
Nucleic acids are polymers made up of monomers called nucleotides.[39] Their function is to store, transmit, and express hereditary information.[36] Nucleotides consist of a phosphate group, a five-carbon sugar, and a nitrogenous base. Ribonucleotides, which contain ribose as the sugar, are the monomers of ribonucleic acid (RNA). In contrast, deoxyribonucleotides contain deoxyribose as the sugar and are constitute the monomers of deoxyribonucleic acid (DNA). RNA and DNA also differ with respect to one of their bases.[39] There are two types of bases: purines and pyrimidines.[39] The purines include guanine (G) and adenine (A) whereas the pyrimidines consist of cytosine (C), uracil (U), and thymine (T). Uracil is used in RNA whereas thymine is used in DNA. Taken together, when the different sugar and bases are take into consideration, there are eight distinct nucleotides that can form two types of nucleic acids: DNA (A, G, C, and T) and RNA (A, G, C, and U).[39]
Cells
Cell theory states that cells are the fundamental units of life, that all living things are composed of one or more cells, and that all cells arise from preexisting cells through cell division.[40] Most cells are very small, with diameters ranging from 1 to 100 micrometers and are therefore only visible under a light or electron microscope.[41] There are generally two types of cells: eukaryotic cells, which contain a nucleus, and prokaryotic cells, which do not. Prokaryotes are single-celled organisms such as bacteria, whereas eukaryotes can be single-celled or multicellular. In multicellular organisms, every cell in the organism's body is derived ultimately from a single cell in a fertilized egg.
Cell structure

Every cell is enclosed within a cell membrane that separates its cytoplasm from the extracellular space.[42] A cell membrane consists of a lipid bilayer, including cholesterols that sit between phospholipids to maintain their fluidity at various temperatures. Cell membranes are semipermeable, allowing small molecules such as oxygen, carbon dioxide, and water to pass through while restricting the movement of larger molecules and charged particles such as ions.[43] Cell membranes also contains membrane proteins, including integral membrane proteins that go across the membrane serving as membrane transporters, and peripheral proteins that loosely attach to the outer side of the cell membrane, acting as enzymes shaping the cell.[44] Cell membranes are involved in various cellular processes such as cell adhesion, storing electrical energy, and cell signalling and serve as the attachment surface for several extracellular structures such as a cell wall, glycocalyx, and cytoskeleton.

Within the cytoplasm of a cell, there are many biomolecules such as proteins and nucleic acids.[45] In addition to biomolecules, eukaryotic cells have specialized structures called organelles that have their own lipid bilayers or are spatially units.[46] These organelles include the cell nucleus, which contains most of the cell's DNA, or mitochondria, which generates adenosine triphosphate (ATP) to power cellular processes. Other organelles such as endoplasmic reticulum and Golgi apparatus play a role in the synthesis and packaging of proteins, respectively. Biomolecules such as proteins can be engulfed by lysosomes, another specialized organelle. Plant cells have additional organelles that distinguish them from animal cells such as a cell wall that provides support for the plant cell, chloroplasts that harvest sunlight energy to produce sugar, and vacuoles that provide storage and structural support as well as being involved in reproduction and breakdown of plant seeds.[46] Eukaryotic cells also have cytoskeleton that is made up of microtubules, intermediate filaments, and microfilaments, all of which provide support for the cell and are involved in the movement of the cell and its organelles.[46] In terms of their structural composition, the microtubules are made up of tubulin (e.g., α-tubulin and β-tubulin whereas intermediate filaments are made up of fibrous proteins.[46] Microfilaments are made up of actin molecules that interact with other strands of proteins.[46]
Metabolism

All cells require energy to sustain cellular processes. Energy is the capacity to do work, which, in thermodynamics, can be calculated using Gibbs free energy. According to the first law of thermodynamics, energy is conserved, i.e., cannot be created or destroyed. Hence, chemical reactions in a cell do not create new energy but are involved instead in the transformation and transfer of energy.[47] Nevertheless, all energy transfers lead to some loss of usable energy, which increases entropy (or state of disorder) as stated by the second law of thermodynamics. As a result, an organism requires continuous input of energy to maintain a low state of entropy. In cells, energy can be transferred as electrons during redox (reduction–oxidation) reactions, stored in covalent bonds, and generated by the movement of ions (e.g., hydrogen, sodium, potassium) across a membrane.
Metabolism is the set of life-sustaining chemical reactions in organisms. The three main purposes of metabolism are: the conversion of food to energy to run cellular processes; the conversion of food/fuel to building blocks for proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments. Metabolic reactions may be categorized as catabolic—the breaking down of compounds (for example, the breaking down of glucose to pyruvate by cellular respiration); or anabolic—the building up (synthesis) of compounds (such as proteins, carbohydrates, lipids, and nucleic acids). Usually, catabolism releases energy, and anabolism consumes energy.
The chemical reactions of metabolism are organized into metabolic pathways, in which one chemical is transformed through a series of steps into another chemical, each step being facilitated by a specific enzyme. Enzymes are crucial to metabolism because they allow organisms to drive desirable reactions that require energy that will not occur by themselves, by coupling them to spontaneous reactions that release energy. Enzymes act as catalysts—they allow a reaction to proceed more rapidly without being consumed by it—by reducing the amount of activation energy needed to convert reactants into products. Enzymes also allow the regulation of the rate of a metabolic reaction, for example in response to changes in the cell's environment or to signals from other cells.
Cellular respiration

Cellular respiration is a set of metabolic reactions and processes that take place in the cells of organisms to convert chemical energy from nutrients into adenosine triphosphate (ATP), and then release waste products.[48] The reactions involved in respiration are catabolic reactions, which break large molecules into smaller ones, releasing energy. Respiration is one of the key ways a cell releases chemical energy to fuel cellular activity. The overall reaction occurs in a series of biochemical steps, some of which are redox reactions. Although cellular respiration is technically a combustion reaction, it clearly does not resemble one when it occurs in a cell because of the slow, controlled release of energy from the series of reactions.
Sugar in the form of glucose is the main nutrient used by animal and plant cells in respiration. Cellular respiration involving oxygen is called aerobic respiration, which has four stages: glycolysis, citric acid cycle (or Krebs cycle), electron transport chain, and oxidative phosphorylation.[49] Glycolysis is a metabolic process that occurs in the cytoplasm whereby glucose is converted into two pyruvates, with two net molecules of ATP being produced at the same time.[49] Each pyruvate is then oxidized into acetyl-CoA by the pyruvate dehydrogenase complex, which also generates NADH and carbon dioxide. Acetyl-Coa enters the citric acid cycle, which takes places inside the mitochondrial matrix. At the end of the cycle, the total yield from 1 glucose (or 2 pyruvates) is 6 NADH, 2 FADH2, and 2 ATP molecules. Finally, the next stage is oxidative phosphorylation, which in eukaryotes, occurs in the mitochondrial cristae. Oxidative phosphorylation comprises the electron transport chain, which is a series of four protein complexes that transfer electrons from one complex to another, thereby releasing energy from NADH and FADH2 that is coupled to the pumping of protons (hydrogen ions) across the inner mitochondrial membrane (chemiosmosis), which generates a proton motive force.[49] Energy from the proton motive force drives the enzyme ATP synthase to synthesize more ATPs by phosphorylating ADPs. The transfer of electrons terminates with molecular oxygen being the final electron acceptor.
If oxygen were not present, pyruvate would not be metabolized by cellular respiration but undergoes a process of fermentation. The pyruvate is not transported into the mitochondrion but remains in the cytoplasm, where it is converted to waste products that may be removed from the cell. This serves the purpose of oxidizing the electron carriers so that they can perform glycolysis again and removing the excess pyruvate. Fermentation oxidizes NADH to NAD+ so it can be re-used in glycolysis. In the absence of oxygen, fermentation prevents the buildup of NADH in the cytoplasm and provides NAD+ for glycolysis. This waste product varies depending on the organism. In skeletal muscles, the waste product is lactic acid. This type of fermentation is called lactic acid fermentation. In strenuous exercise, when energy demands exceed energy supply, the respiratory chain cannot process all of the hydrogen atoms joined by NADH. During anaerobic glycolysis, NAD+ regenerates when pairs of hydrogen combine with pyruvate to form lactate. Lactate formation is catalyzed by lactate dehydrogenase in a reversible reaction. Lactate can also be used as an indirect precursor for liver glycogen. During recovery, when oxygen becomes available, NAD+ attaches to hydrogen from lactate to form ATP. In yeast, the waste products are ethanol and carbon dioxide. This type of fermentation is known as alcoholic or ethanol fermentation. The ATP generated in this process is made by substrate-level phosphorylation, which does not require oxygen.
Photosynthesis

Photosynthesis is a process used by plants and other organisms to convert light energy into chemical energy that can later be released to fuel the organism's metabolic activities via cellular respiration. This chemical energy is stored in carbohydrate molecules, such as sugars, which are synthesized from carbon dioxide and water.[50][51][52] In most cases, oxygen is also released as a waste product. Most plants, algae, and cyanobacteria perform photosynthesis, which is largely responsible for producing and maintaining the oxygen content of the Earth's atmosphere, and supplies most of the energy necessary for life on Earth.[53]
Photosynthesis has four stages: Light absorption, electron transport, ATP synthesis, and carbon fixation.[49] Light absorption is the initial step of photosynthesis whereby light energy is absorbed by chlorophyll pigments attached to proteins in the thylakoid membranes. The absorbed light energy is used to remove electrons from a donor (water) to a primary electron acceptor, a quinone designated as Q. In the second stage, electrons move from the quinone primary electron acceptor through a series of electron carriers until they reach a final electron acceptor, which is usually the oxidized form of NADP+, which is reduced to NADPH, a process that takes place in a protein complex called photosystem I (PSI). The transport of electrons is coupled to the movement of protons (or hydrogen) from the stroma to the thylakoid membrane, which forms a pH gradient across the membrane as hydrogen becomes more concentrated in the lumen than in the stroma. This is analogous to the proton-motive force generated across the inner mitochondrial membrane in aerobic respiration.[49]
During the third stage of photosynthesis, the movement of protons down their concentration gradients from the thylakoid lumen to the stroma through the ATP synthase is coupled to the synthesis of ATP by that same ATP synthase.[49] The NADPH and ATPs generated by the light-dependent reactions in the second and third stages, respectively, provide the energy and electrons to drive the synthesis of glucose by fixing atmospheric carbon dioxide into existing organic carbon compounds, such as ribulose bisphosphate (RuBP) in a sequence of light-independent (or dark) reactions called the Calvin cycle.[54]
Cell signaling
Cell signaling (or communication) is the ability of cells to receive, process, and transmit signals with its environment and with itself.[55][56] Signals can be non-chemical such as light, electrical impulses, and heat, or chemical signals (or ligands) that interact with receptors, which can be found embedded in the cell membrane of another cell or located deep inside a cell.[57][56] There are generally four types of chemical signals: autocrine, paracrine, juxtacrine, and hormones.[57] In autocrine signaling, the ligand affects the same cell that releases it. Tumor cells, for example, can reproduce uncontrollably because they release signals that initiate their own self-division. In paracrine signaling, the ligand diffuses to nearby cells and affects them. For example, brain cells called neurons release ligands called neurotransmitters that diffuse across a synaptic cleft to bind with a receptor on an adjacent cell such as another neuron or muscle cell. In juxtacrine signaling, there is direct contact between the signaling and responding cells. Finally, hormones are ligands that travel through the circulatory systems of animals or vascular systems of plants to reach their target cells. Once a ligand binds with a receptor, it can influence the behavior of another cell, depending on the type of receptor. For instance, neurotransmitters that bind with an inotropic receptor can alter the excitability of a target cell. Other types of receptors include protein kinase receptors (e.g., receptor for the hormone insulin) and G protein-coupled receptors. Activation of G protein-coupled receptors can initiate second messenger cascades. The process by which a chemical or physical signal is transmitted through a cell as a series of molecular events is called signal transduction
Cell cycle

The cell cycle is a series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA and some of its organelles, and the subsequent partitioning of its cytoplasm into two daughter cells in a process called cell division.[58] In eukaryotes (i.e., animal, plant, fungal, and protist cells), there are two distinct types of cell division: mitosis and meiosis.[59] Mitosis is part of the cell cycle, in which replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis (division of the nucleus) is preceded by the S stage of interphase (during which the DNA is replicated) and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis all together define the mitotic phase of an animal cell cycle—the division of the mother cell into two genetically identical daughter cells.[60] The cell cycle is a vital process by which a single-celled fertilized egg develops into a mature organism, as well as the process by which hair, skin, blood cells, and some internal organs are renewed. After cell division, each of the daughter cells begin the interphase of a new cycle. In contrast to mitosis, meiosis results in four haploid daughter cells by undergoing one round of DNA replication followed by two divisions.[61] Homologous chromosomes are separated in the first division (meiosis I), and sister chromatids are separated in the second division (meiosis II). Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.
Prokaryotes (i.e., archaea and bacteria) can also undergo cell division (or binary fission). Unlike the processes of mitosis and meiosis in eukaryotes, binary fission takes in prokaryotes takes place without the formation of a spindle apparatus on the cell. Before binary fission, DNA in the bacterium is tightly coiled. After it has uncoiled and duplicated, it is pulled to the separate poles of the bacterium as it increases the size to prepare for splitting. Growth of a new cell wall begins to separate the bacterium (triggered by FtsZ polymerization and "Z-ring" formation)[62] The new cell wall (septum) fully develops, resulting in the complete split of the bacterium. The new daughter cells have tightly coiled DNA rods, ribosomes, and plasmids.
Genetics
Inheritance

Genetics is the scientific study of inheritance.[63][64][65] Mendelian inheritance, specifically, is the process by which genes and traits are passed on from parents to offspring.[28] It was formulated by Gregor Mendel, based on his work with pea plants in the mid-nineteenth century. Mendel established several principles of inheritance. The first is that genetic characteristics, which are now called alleles, are discrete and have alternate forms (e.g., purple vs. white or tall vs. dwarf), each inherited from one of two parents. Based on his law of dominance and uniformity, which states that some alleles are dominant while others are recessive; an organism with at least one dominant allele will display the phenotype of that dominant allele.[66] Exceptions to this rule include penetrance and expressivity.[28] Mendel noted that during gamete formation, the alleles for each gene segregate from each other so that each gamete carries only one allele for each gene, which is stated by his law of segregation. Heterozygotic individuals produce gametes with an equal frequency of two alleles. Finally, Mendel formulated the law of independent assortment, which states that genes of different traits can segregate independently during the formation of gametes, i.e., genes are unlinked. An exception to this rule would include traits that are sex-linked. Test crosses can be performed to experimentally determine the underlying genotype of an organism with a dominant phenotype.[67] A Punnett square can be used to predict the results of a test cross. The chromosome theory of inheritance, which states that genes are found on chromosomes, was supported by Thomas Morgans's experiments with fruit flies, which established the sex linkage between eye color and sex in these insects.[68] In humans and other mammals (e.g., dogs), it is not feasible or practical to conduct test cross experiments. Instead, pedigrees, which are genetic representations of family trees,[69] are used instead to trace the inheritance of a specific trait or disease through multiple generations.[70]
DNA

A gene is a unit of heredity that corresponds to a region of deoxyribonucleic acid (DNA) that carries genetic information that influences the form or function of an organism in specific ways. DNA is a molecule composed of two polynucleotide chains that coil around each other to form a double helix, which was first described by James Watson and Francis Crick in 1953.[71] It is found as linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. A chromosome is an organized structure consisting of DNA and histones. The set of chromosomes in a cell and any other hereditary information found in the mitochondria, chloroplasts, or other locations is collectively known as a cell's genome. In eukaryotes, genomic DNA is localized in the cell nucleus, or with small amounts in mitochondria and chloroplasts.[72] In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid.[73] The genetic information in a genome is held within genes, and the complete assemblage of this information in an organism is called its genotype.[74] Genes encode the information needed by cells for the synthesis of proteins, which in turn play a central role in influencing the final phenotype of the organism.
The two polynucleotide strands that make up DNA run in opposite directions to each other and are thus antiparallel. Each strand is composed of nucleotides,[75][76] with each nucleotide containing one of four nitrogenous bases (cytosine [C], guanine [G], adenine [A] or thymine [T]), a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. It is the sequence of these four bases along the backbone that encodes genetic information. Bases of the two polynucleotide strands are bound together by hydrogen bonds, according to base pairing rules (A with T and C with G), to make double-stranded DNA. The bases are divided into two groups: pyrimidines and purines. In DNA, the pyrimidines are thymine and cytosine whereas the purines are adenine and guanine.
There are grooves that run along the entire length of the double helix due to the uneven spacing of the DNA strands relative to each other.[71] Both grooves differ in size, with the major groove being larger and therefore more accessible to the binding of proteins than the minor groove.[71] The outer edges of the bases are exposed to these grooves and are therefore accessible for additional hydrogen bonding.[71] Because each groove can have two possible base-pair configurations (G-C and A-T), there are four possible base-pair configurations within the entire double helix, each of which is chemically distinct from another.[71] As a result, protein molecules are able to recognize and bind to specific base-pair sequences, which is the basis of specific DNA-protein interactions.
DNA replication is a semiconservative process whereby each strand serves as a template for a new strand of DNA.[71] The process begins with the unwounding of the double helix at an origin of replication, which separates the two strands, thereby making them available as two templates. This is then followed by the binding of the enzyme primase to the template to synthesize a starter RNA (or DNA in some viruses) strand called a primer from the 5' to 3' location.[71] Once the primer is completed, the primase is released from the template, followed by the binding of the enzyme DNA polymerase to the same template to synthesize new DNA. The rate of DNA replication in a living cell was measured as 749 nucleotides added per second under ideal conditions.[77]
DNA replication is not perfect as the DNA polymerase sometimes insert bases that are not complementary to the template (e.g., putting in A in the strand opposite to G in the template strand).[71] In eukaryotes, the initial error or mutation rate is about 1 in 100,000.[71] Proofreading and mismatch repair are the two mechanisms that repair these errors, which reduces the mutation rate to 10−10, particularly before and after a cell cycle.[71]
Mutations are heritable changes in DNA.[71] They can arise spontaneously as a result of replication errors that were not corrected by proofreading or can be induced by an environmental mutagen such as a chemical (e.g., nitrous acid, benzopyrene) or radiation (e.g., x-ray, gamma ray, ultraviolet radiation, particles emitted by unstable isotopes).[71] Mutations can appear as a change in single base or at a larger scale involving chromosomal mutations such as deletions, inversions, or translocations.[71]
In multicellular organisms, mutations can occur in somatic or germline cells.[71] In somatic cells, the mutations are passed on to daughter cells during mitosis.[71] In a germline cell such as a sperm or an egg, the mutation will appear in an organism at fertilization.[71] Mutations can lead to several types of phenotypic effects such as silent, loss-of-function, gain-of-function, and conditional mutations.[71]
Some mutations can be beneficial, as they are a source of genetic variation for evolution.[71] Others can be harmful if they were to result in a loss of function of genes needed for survival.[71] Mutagens such as carcinogens are typically avoided as a matter of public health policy goals.[71] One example is the banning of chlorofluorocarbons (CFC) by the Montreal Protocol, as CFCs tend to deplete the ozone layer, resulting in more ultraviolet radiation from the sun passing through the Earth's upper atmosphere, thereby causing somatic mutations that can lead to skin cancer.[71] Similarly, smoking bans have been enforced throughout the world in an effort to reduce the incidence of lung cancer.[71]
Gene expression

Gene expression is the molecular process by which a genotype gives rise to a phenotype, i.e., observable trait. The genetic information stored in DNA represents the genotype, whereas the phenotype results from the synthesis of proteins that control an organism's structure and development, or that act as enzymes catalyzing specific metabolic pathways. This process is summarized by the central dogma of molecular biology, which was formulated by Francis Crick in 1958.[78][79][80] According to the Central Dogma, genetic information flows from DNA to RNA to protein. Hence, there are two gene expression processes: transcription (DNA to RNA) and translation (RNA to protein).[81] These processes are used by all life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and are exploited by viruses—to generate the macromolecular machinery for life.
During transcription, messenger RNA (mRNA) strands are created using DNA strands as a template, which is initiated when RNA polymerase binds to a DNA sequence called a promoter, which instructs the RNA to begin transcription of one of the two DNA strands.[82] The DNA bases are exchanged for their corresponding bases except in the case of thymine (T), for which RNA substitutes uracil (U).[83] In eukaryotes, a large part of DNA (e.g., >98% in humans) contain non-coding called introns, which do not serve as patterns for protein sequences. The coding regions or exons are interspersed along with the introns in the primary transcript (or pre-mRNA).[82] Before translation, the pre-mRNA undergoes further processing whereby the introns are removed (or spliced out), leaving only the spliced exons in the mature mRNA strand.[82]
The translation of mRNA to protein occurs in ribosomes, whereby the transcribed mRNA strand specifies the sequence of amino acids within proteins using the genetic code. Gene products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA.[84][85]
Gene regulation

The regulation of gene expression (or gene regulation) by environmental factors and during different stages of development can occur at each step of the process such as transcription, RNA splicing, translation, and post-translational modification of a protein.[86]
The ability of gene transcription to be regulated allows for the conservation of energy as cells will only make proteins when needed.[86] Gene expression can be influenced by positive or negative regulation, depending on which of the two types of regulatory proteins called transcription factors bind to the DNA sequence close to or at a promoter.[86] A cluster of genes that share the same promoter is called an operon, found mainly in prokaryotes and some lower eukaryotes (e.g., Caenorhabditis elegans).[86][87] It was first identified in Escherichia coli—a prokaryotic cell that can be found in the intestines of humans and other animals—in the 1960s by François Jacob and Jacques Monod.[86] They studied the prokaryotic cell's lac operon, which is part of three genes (lacZ, lacY, and lacA) that encode three lactose-metabolizing enzymes (β-galactosidase, β-galactoside permease, and β-galactoside transacetylase).[86] In positive regulation of gene expression, the activator is the transcription factor that stimulates transcription when it binds to the sequence near or at the promoter. In contrast, negative regulation occurs when another transcription factor called a repressor binds to a DNA sequence called an operator, which is part of an operon, to prevent transcription. When a repressor binds to a repressible operon (e.g., trp operon), it does so only in the presence of a corepressor. Repressors can be inhibited by compounds called inducers (e.g., allolactose), which exert their effects by binding to a repressor to prevent it from binding to an operator, thereby allowing transcription to occur.[86] Specific genes that can be activated by inducers are called inducible genes (e.g., lacZ or lacA in E. coli), which are in contrast to constitutive genes that are almost always active.[86] In contrast to both, structural genes encode proteins that are not involved in gene regulation.[86]
In prokaryotic cells, transcription is regulated by proteins called sigma factors, which bind to RNA polymerase and direct it to specific promoters.[86] Similarly, transcription factors in eukaryotic cells can also coordinate the expression of a group of genes, even if the genes themselves are located on different chromosomes.[86] Coordination of these genes can occur as long as they share the same regulatory DNA sequence that bind to the same transcription factors.[86] Promoters in eukaryotic cells are more diverse but tend to contain a core sequence that RNA polymerase can bind to, with the most common sequence being the TATA box, which contains multiple repeating A and T bases.[86] Specifically, RNA polymerase II is the RNA polymerase that binds to a promoter to initiate transcription of protein-coding genes in eukaryotes, but only in the presence of multiple general transcription factors, which are distinct from the transcription factors that have regulatory effects, i.e., activators and repressors.[86] In eukaryotic cells, DNA sequences that bind with activators are called enhances whereas those sequences that bind with repressors are called silencers.[86] Transcription factors such as nuclear factor of activated T-cells (NFAT) are able to identify specific nucleotide sequence based on the base sequence (e.g., CGAGGAAAATTG for NFAT) of the binding site, which determines the arrangement of the chemical groups within that sequence that allows for specific DNA-protein interactions.[86] The expression of transcription factors is what underlies cellular differentiation in a developing embryo.[86]
In addition to regulatory events involving the promoter, gene expression can also be regulated by epigenetic changes to chromatin, which is a complex of DNA and protein found in eukaryotic cells.[86]
Post-transcriptional control of mRNA can involve the alternative splicing of primary mRNA transcripts, resulting in a single gene giving rise to different mature mRNAs that encode a family of different proteins.[86][88] A well-studied example is the Sxl gene in Drosophila, which determines the sex in these animals. The gene itself contains four exons and alternative splicing of its pre-mRNA transcript can generate two active forms of the Sxl protein in female flies and one in inactive form of the protein in males.[86] Another example is the human immunodeficiency virus (HIV), which has a single pre-mRNA transcript that can generate up to nine proteins as a result of alternative splicing.[86] In humans, eighty percent of all 21,000 genes are alternatively spliced.[86] Given that both chimpanzees and humans have a similar number of genes, it is thought that alternative splicing might have contributed to the latter's complexity due to the greater number of alternative splicing in the human brain than in the brain of chimpanzees.[86]
Translation can be regulated in three known ways, one of which involves the binding of tiny RNA molecules called microRNA (miRNA) to a target mRNA transcript, which inhibits its translation and causes it to degrade.[86] Translation can also be inhibited by the modification of the 5' cap by substituting the modified guanosine triphosphate (GTP) at the 5' end of an mRNA for an unmodified GTP molecule.[86] Finally, translational repressor proteins can bind to mRNAs and prevent them from attaching to a ribosome, thereby blocking translation.[86]
Once translated, the stability of proteins can be regulated by being targeted for degradation.[86] A common example is when an enzyme attaches a regulatory protein called ubiquitin to the lysine residue of a targeted protein.[86] Other ubiquitins then attached to the primary ubiquitin to form a polyubiquitinated protein, which then enters a much larger protein complex called proteasome.[86] Once the polyubiquitinated protein enters the proteasome, the polyubiquitin detaches from the target protein, which is unfolded by the proteasome in an ATP-dependent manner, allowing it to be hydrolyzed by three proteases.[86]
Genomes

A genome is an organism's complete set of DNA, including all of its genes.[89] Sequencing and analysis of genomes can be done using high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes.[90][91][92] The genomes of prokaryotes are small, compact, and diverse. In contrast, the genomes of eukaryotes are larger and more complex such as having more regulatory sequences and much of its genome are made up of non-coding DNA sequences for functional RNA (rRNA, tRNA, and mRNA) or regulatory sequences. The genomes of various model organisms such as arabidopsis, fruit fly, mice, nematodes, and yeast have been sequenced. The Human Genome Project was a major undertaking by the international scientific community to sequence the entire human genome, which was completed in 2003.[93] The sequencing of the human genome has yielded practical applications such as DNA fingerprinting, which can be used for paternity testing and forensics. In medicine, sequencing of the entire human genome has allowed for the identification of mutations that cause tumors as well as genes that cause a specific genetic disorder.[93] The sequencing of genomes from various organisms has led to the emergence of comparative genomics, which aims to draw comparisons of genes from the genomes of those different organisms.[93]
Many genes encode more than one protein, with posttranslational modifications increasing the diversity of proteins within a cell. An organism's proteome is its entire set of proteins expressed by its genome and proteomics seeks to study the complete set of proteins produced by an organism.[93] Because many proteins are enzymes, their activities tend to affects the concentrations of substrates and products. Thus, as the proteome changes, so do the amount of small molecules or metabolites.[93] The complete set of small molecules in a cell or organism is called a metabolome and metabolomics is the study of the metabolome in relation to the physiological activity of a cell or organism.[93]
Biotechnology

Biotechnology is the use of cells or organisms to develop products for humans.[94] One commonly used technology with wide applications is the creation of recombinant DNA, which is a DNA molecule assembled from two or more sources in a laboratory. Before the advent of polymerase chain reaction, biologists would manipulate DNA by cutting it into smaller fragments using restriction enzymes. They would then purify and analyze the fragments using gel electrophoresis and then later recombine the fragments into a novel DNA sequence using DNA ligase.[94] The recombinant DNA is then cloned by inserting it into a host cell, a process known as transformation if the host cells were bacteria such as E. coli, or transfection if the host cells were eukaryotic cells like yeast, plant, or animal cells. Once the host cell or organism has received and integrated the recombinant DNA, it is described as transgenic.[94]
A recombinant DNA can be inserted in one of two ways. A common method is to simply insert the DNA into a host chromosome, with the site of insertion being random.[94] Another approach would be to insert the recombinant DNA as part of another DNA sequence called a vector, which then integrates into the host chromosome or has its own origin of DNA replication, thereby allowing to replicate independently of the host chromosome.[94] Plasmids from bacterial cells such as E. coli are typically used as vectors due to their relatively small size (e.g. 2000–6000 base pairs in E. coli), presence of restriction enzymes, genes that are resistant to antibiotics, and the presence of an origin of replication.[94] A gene coding for a selectable marker such as antibiotic resistance is also incorporated into the vector.[94] Inclusion of this market allows for the selection of only those host cells that contained the recombinant DNA while discarding those that do not.[94] Moreover, the marker also serves as a reporter gene that once expressed, can be easily detected and measured.[94]
Once the recombinant DNA is inside individual bacterial cells, those cells are then plated and allowed to grow into a colony that contains millions of transgenic cells that carry the same recombinant DNA.[95] These transgenic cells then produce large quantities of the transgene product such as human insulin, which was the first medicine to be made using recombinant DNA technology.[94]
One of the goals of molecular cloning is to identify the function of specific DNA sequences and the proteins they encode.[94] For a specific DNA sequence to be studied and manipulated, millions of copies of DNA fragments containing that DNA sequence need to be made.[94] This involves breaking down an intact genome, which is much too large to be introduced into a host cell, into smaller DNA fragments. Although no longer intact, the collection of these DNA fragments still make up an organism's genome, with the collection itself being referred to as a genomic library, due to the ability to search and retrieve specific DNA fragments for further study, analogous to the process of retrieving a book from a regular library.[94] DNA fragments can be obtained using restriction enzymes and other processes such as mechanical shearing. Each obtained fragment is then inserted into a vector that is taken up by a bacterial host cell. The host cell is then allowed to proliferate on a selective medium (e.g., antibiotic resistance), which produces a colony of these recombinant cells, each of which contains many copies of the same DNA fragment.[94] These colonies can be grown by spreading them over a solid medium in Petri dishes, which are incubated at a suitable temperature. One dish alone can hold thousands of bacterial colonies, which can be easily screened for a specific DNA sequence.[94] The sequence can be identified by first duplicating a Petri dish with bacterial colonies and then exposing the DNA of the duplicated colonies for hybridization, which involves labeling them with complementary radioactive or fluorescent nucleotides.[94]
Smaller DNA libraries that contain genes from a specific tissue can be created using complementary DNA (cDNA).[94] The collection of these cDNAs from a specific tissue at a particular time is called a cDNA library, which provides a "snapshot" of transcription patterns of cells at a specific location and time.[94]
Other biotechnology tools include DNA microarrays, expression vectors, synthetic genomics, and CRISPR gene editing.[94][96] Other approaches such as pharming can produce large quantities of medically useful products through the use of genetically modified organisms.[94] Many of these other tools also have wide applications such as creating medically useful proteins, or improving plant cultivation and animal husbandry.[94]
Genes, development, and evolution
Development is the process by which a multicellular organism (plant or animal) goes through a series of a changes, starting from a single cell, and taking on various forms that are characteristic of its life cycle.[98] There are four key processes that underlie development: Determination, differentiation, morphogenesis, and growth. Determination sets the developmental fate of a cell, which becomes more restrictive during development. Differentiation is the process by which specialized cells from less specialized cells such as stem cells.[99][100] Stem cells are undifferentiated or partially differentiated cells that can differentiate into various types of cells and proliferate indefinitely to produce more of the same stem cell.[101] Cellular differentiation dramatically changes a cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals, which are largely due to highly controlled modifications in gene expression and epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the DNA sequence itself.[102] Thus, different cells can have very different physical characteristics despite having the same genome. Morphogenesis, or the development of body form, is the result of spatial differences in gene expression.[98] Specially, the organization of differentiated tissues into specific structures such as arms or wings, which is known as pattern formation, is governed by morphogens, signaling molecules that move from one group of cells to surrounding cells, creating a morphogen gradient as described by the French flag model. Apoptosis, or programmed cell death, also occurs during morphogenesis, such as the death of cells between digits in human embryonic development, which frees up individual fingers and toes. Expression of transcription factor genes can determine organ placement in a plant and a cascade of transcription factors themselves can establish body segmentation in a fruit fly.[98]
A small fraction of the genes in an organism's genome called the developmental-genetic toolkit control the development of that organism. These toolkit genes are highly conserved among phyla, meaning that they are ancient and very similar in widely separated groups of animals. Differences in deployment of toolkit genes affect the body plan and the number, identity, and pattern of body parts. Among the most important toolkit genes are the Hox genes. Hox genes determine where repeating parts, such as the many vertebrae of snakes, will grow in a developing embryo or larva.[103] Variations in the toolkit may have produced a large part of the morphological evolution of animals. The toolkit can drive evolution in two ways. A toolkit gene can be expressed in a different pattern, as when the beak of Darwin's large ground-finch was enlarged by the BMP gene,[104] or when snakes lost their legs as Distal-less (Dlx) genes became under-expressed or not expressed at all in the places where other reptiles continued to form their limbs.[105] Or, a toolkit gene can acquire a new function, as seen in the many functions of that same gene, distal-less, which controls such diverse structures as the mandible in vertebrates,[106][107] legs and antennae in the fruit fly,[108] and eyespot pattern in butterfly wings.[109] Given that small changes in toolbox genes can cause significant changes in body structures, they have often enabled convergent or parallel evolution.
Evolution
Evolutionary processes

A central organizing concept in biology is that life changes and develops through evolution, which is the change in heritable characteristics of populations over successive generations.[110][111] Evolution is now used to explain the great variations of life on Earth. The term evolution was introduced into the scientific lexicon by Jean-Baptiste de Lamarck in 1809.[112][113] He proposed that evolution occurred as a result of inheritance of acquired characteristics, which was unconvincing but there were no alternative explanations at the time.[112] Charles Darwin, an English naturalist, had returned to England in 1836 from his five-year travels on the HMS Beagle where he studied rocks and collected plants and animals from various parts of the world such as the Galápagos Islands.[112] He had also read Principles of Geology by Charles Lyell and An Essay on the Principle of Population by Thomas Malthus and was influenced by them.[114] Based on his observations and readings, Darwin began to formulate his theory of evolution by natural selection to explain the diversity of plants and animals in different parts of the world.[112][114] Alfred Russel Wallace, another English naturalist who had studied plants and animals in the Malay Archipelago, also came to the same idea, but later and independently of Darwin.[112] Both Darwin and Wallace jointly presented their essay and manuscript, respectively, at the Linnaean Society of London in 1858, giving them both credit for their discovery of evolution by natural selection.[112][115][116][117][118] Darwin would later publish his book On the Origin of Species in 1859, which explained in detail how the process of evolution by natural selection works.[112]
To explain natural selection, Darwin drew an analogy with humans modifying animals through artificial selection, whereby animals were selectively bred for specific traits, which has given rise to individuals that no longer resemble their wild ancestors.[114] Darwin argued that in the natural world, it was nature that played the role of humans in selecting for specific traits. He came to this conclusion based on two observations and two inferences.[114] First, members of any population tend to vary with respect to their heritable traits. Second, all species tend to produce more offspring than can be supported by their respective environments, resulting in many individuals not surviving and reproducing.[114] Based on these observations, Darwin inferred that those individuals who possessed heritable traits that are better adapted to their environments are more likely to survive and produce more offspring than other individuals.[114] He further inferred that the unequal or differential survival and reproduction of certain individuals over others will lead to the accumulation of favorable traits over successive generations, thereby increasing the match between the organisms and their environment.[114][119][120] Thus, taken together, natural selection is the differential survival and reproduction of individuals in subsequent generations due to differences in or more heritable traits.[121][114][112]
Darwin was not aware of Mendel's work of inheritance and so the exact mechanism of inheritance that underlie natural selection was not well-understood[122] until the early 20th century when the modern synthesis reconciled Darwinian evolution with classical genetics, which established a neo-Darwinian perspective of evolution by natural selection.[121] This perspective holds that evolution occurs when there are changes in the allele frequencies within a population of interbreeding organisms. In the absence of any evolutionary process acting on a large random mating population, the allele frequencies will remain constant across generations as described by the Hardy–Weinberg principle.[123]
Another process that drives evolution is genetic drift, which is the random fluctuations of allele frequencies within a population from one generation to the next.[124] When selective forces are absent or relatively weak, allele frequencies are equally likely to drift upward or downward at each successive generation because the alleles are subject to sampling error.[125] This drift halts when an allele eventually becomes fixed, either by disappearing from the population or replacing the other alleles entirely. Genetic drift may therefore eliminate some alleles from a population due to chance alone.
Speciation

A species is a group of organisms that mate with one another and speciation is the process by which one lineage splits into two lineages as a result of having evolved independently from each other.[126] For speciation to occur, there has to be reproductive isolation.[126] Reproductive isolation can result from incompatibilities between genes as described by Bateson–Dobzhansky–Muller model. Reproductive isolation also tends to increase with genetic divergence. Speciation can occur when there are physical barriers that divide an ancestral species, a process known as allopatric speciation.[126] In contrast, sympatric speciation occurs in the absence of physical barriers.
Pre-zygotic isolation such as mechanical, temporal, behavioral, habitat, and gametic isolations can prevent different species from hybridizing.[126] Similarly, post-zygotic isolations can result in hybridization being selected against due to the lower viability of hybrids or hybrid infertility (e.g., mule). Hybrid zones can emerge if there were to be incomplete reproductive isolation between two closely related species.
Phylogeny
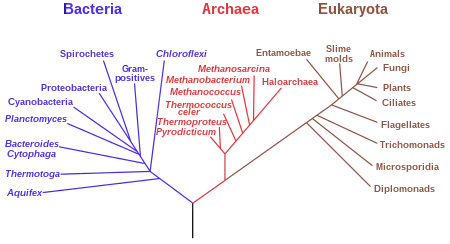
A phylogeny is an evolutionary history of a specific group of organisms or their genes.[127] It can be represented using a phylogenetic tree, which is a diagram showing lines of descent among organisms or their genes. Each line drawn on the time axis of a tree represents a lineage of descendants of a particular species or population. When a lineage divides into two, it is represented as a node (or split) on the phylogenetic tree. The more splits there are over time, the more branches there will be on the tree, with the common ancestor of all the organisms in that tree being represented by the root of that tree. Phylogenetic trees may portray the evolutionary history of all life forms, a major evolutionary group (e.g., insects), or an even smaller group of closely related species. Within a tree, any group of species designated by a name is a taxon (e.g., humans, primates, mammals, or vertebrates) and a taxon that consists of all its evolutionary descendants is a clade, otherwise known as a monophyletic taxon.[127] Closely related species are referred to as sister species and closely related clades are sister clades. In contrast to a monophyletic group, a polyphyletic group does not include its common ancestor whereas a paraphyletic group does not include all the descendants of a common ancestor.[127]
Phylogenetic trees are the basis for comparing and grouping different species.[127] Different species that share a feature inherited from a common ancestor are described as having homologous features (or synapomorphy).[128][129][127] Homologous features may be any heritable traits such as DNA sequence, protein structures, anatomical features, and behavior patterns. A vertebral column is an example of a homologous feature shared by all vertebrate animals. Traits that have a similar form or function but were not derived from a common ancestor are described as analogous features. Phylogenies can be reconstructed for a group of organisms of primary interests, which are called the ingroup. A species or group that is closely related to the ingroup but is phylogenetically outside of it is called the outgroup, which serves a reference point in the tree. The root of the tree is located between the ingroup and the outgroup.[127] When phylogenetic trees are reconstructed, multiple trees with different evolutionary histories can be generated. Based on the principle of Parsimony (or Occam's razor), the tree that is favored is the one with the fewest evolutionary changes needed to be assumed over all traits in all groups. Computational algorithms can be used to determine how a tree might have evolved given the evidence.[127]
Phylogeny provides the basis of biological classification, which is based on Linnaean taxonomy that was developed by Carl Linnaeus in the 18th century.[127] This classification system is rank-based, with the highest rank being the domain followed by kingdom, phylum, class, order, family, genus, and species.[127] All organisms can be classified as belonging to one of three domains: Archaea (originally Archaebacteria); bacteria (originally eubacteria), or eukarya (includes the protist, fungi, plant, and animal kingdoms).[130] A binomial nomenclature is used to classify different species. Based on this system, each species is given two names, one for its genus and another for its species.[127] For example, humans are Homo sapiens, with Homo being the genus and sapiens being the species. By convention, the scientific names of organisms are italicized, with only the first letter of the genus capitalized.[131][132]
History of life
Life timeline | ||||||||||||||||||||||||||||||||||||||||||||||
−4500 — – — – −4000 — – — – −3500 — – — – −3000 — – — – −2500 — – — – −2000 — – — – −1500 — – — – −1000 — – — – −500 — – — – 0 — | Water Single-celled life Photosynthesis H a d e a n |
| ||||||||||||||||||||||||||||||||||||||||||||
(million years ago) *Ice Ages | ||||||||||||||||||||||||||||||||||||||||||||||
The history of life on Earth traces the processes by which organisms have evolved from the earliest emergence of life to present day. Earth formed about 4.5 billion years ago and all life on Earth, both living and extinct, descended from a last universal common ancestor that lived about 3.5 billion years ago.[133][134] The dating of the Earth's history can be done using several geological methods such as stratigraphy, radiometric dating, and paleomagnetic dating.[135] Based on these methods, geologists have developed a geologic time scale that divides the history of the Earth into major divisions, starting with four eons (Hadean, Archean, Proterozoic, and Phanerozoic), the first three of which are collectively known as the Precambrian, which lasted approximately 4 billion years.[135] Each eon can be divided into eras, with the Phanerozoic eon that began 539 million years ago[136] being subdivided into Paleozoic, Mesozoic, and Cenozoic eras.[135] These three eras together comprise eleven periods (Cambrian, Ordovician, Silurian, Devonian, Carboniferous, Permian, Triassic, Jurassic, Cretaceous, Tertiary, and Quaternary) and each period into epochs.[135]
The similarities among all known present-day species indicate that they have diverged through the process of evolution from their common ancestor.[137] Biologists regard the ubiquity of the genetic code as evidence of universal common descent for all bacteria, archaea, and eukaryotes.[138][10][139][140] Microbal mats of coexisting bacteria and archaea were the dominant form of life in the early Archean epoch and many of the major steps in early evolution are thought to have taken place in this environment.[141] The earliest evidence of eukaryotes dates from 1.85 billion years ago,[142][143] and while they may have been present earlier, their diversification accelerated when they started using oxygen in their metabolism. Later, around 1.7 billion years ago, multicellular organisms began to appear, with differentiated cells performing specialised functions.[144]
Algae-like multicellular land plants are dated back even to about 1 billion years ago,[145] although evidence suggests that microorganisms formed the earliest terrestrial ecosystems, at least 2.7 billion years ago.[146] Microorganisms are thought to have paved the way for the inception of land plants in the Ordovician period. Land plants were so successful that they are thought to have contributed to the Late Devonian extinction event.[147]
Ediacara biota appear during the Ediacaran period,[148] while vertebrates, along with most other modern phyla originated about 525 million years ago during the Cambrian explosion.[149] During the Permian period, synapsids, including the ancestors of mammals, dominated the land,[150] but most of this group became extinct in the Permian–Triassic extinction event 252 million years ago.[151] During the recovery from this catastrophe, archosaurs became the most abundant land vertebrates;[152] one archosaur group, the dinosaurs, dominated the Jurassic and Cretaceous periods.[153] After the Cretaceous–Paleogene extinction event 66 million years ago killed off the non-avian dinosaurs,[154] mammals increased rapidly in size and diversity.[155] Such mass extinctions may have accelerated evolution by providing opportunities for new groups of organisms to diversify.[156]
Diversity
Bacteria and Archaea

Bacteria are a type of cell that constitute a large domain of prokaryotic microorganisms. Typically a few micrometers in length, bacteria have a number of shapes, ranging from spheres to rods and spirals. Bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste,[157] and the deep biosphere of the earth's crust. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised, and only about 27 percent of the bacterial phyla have species that can be grown in the laboratory.[158]
Archaea constitute the other domain of prokaryotic cells and were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), a term that has fallen out of use.[159] Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat and square cells of Haloquadratum walsbyi.[160] Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved in transcription and translation. Other aspects of archaeal biochemistry are unique, such as their reliance on ether lipids in their cell membranes,[161] including archaeols. Archaea use more energy sources than eukaryotes: these range from organic compounds, such as sugars, to ammonia, metal ions or even hydrogen gas. Salt-tolerant archaea (the Haloarchaea) use sunlight as an energy source, and other species of archaea fix carbon, but unlike plants and cyanobacteria, no known species of archaea does both. Archaea reproduce asexually by binary fission, fragmentation, or budding; unlike bacteria, no known species of Archaea form endospores.
The first observed archaea were extremophiles, living in extreme environments, such as hot springs and salt lakes with no other organisms. Improved molecular detection tools led to the discovery of archaea in almost every habitat, including soil, oceans, and marshlands. Archaea are particularly numerous in the oceans, and the archaea in plankton may be one of the most abundant groups of organisms on the planet.
Archaea are a major part of Earth's life. They are part of the microbiota of all organisms. In the human microbiome, they are important in the gut, mouth, and on the skin.[162] Their morphological, metabolic, and geographical diversity permits them to play multiple ecological roles: carbon fixation; nitrogen cycling; organic compound turnover; and maintaining microbial symbiotic and syntrophic communities, for example.[163]
Protists

Eukaryotes are hypothesized to have split from archaea, which was followed by their endosymbioses with bacteria (or symbiogenesis) that gave rise to mitochondria and chloroplasts, both of which are now part of modern-day eukaryotic cells.[164] The major lineages of eukaryotes diversified in the Precambrian about 1.5 billion years ago and can be classified into eight major clades: alveolates, excavates, stramenopiles, plants, rhizarians, amoebozoans, fungi, and animals.[164] Five of these clades are collectively known as protists, which are mostly microscopic eukaryotic organisms that are not plants, fungi, or animals.[164] While it is likely that protists share a common ancestor (the last eukaryotic common ancestor),[165] protists by themselves do not constitute a separate clade as some protists may be more closely related to plants, fungi, or animals than they are to other protists. Like groupings such as algae, invertebrates, or protozoans, the protist grouping is not a formal taxonomic group but is used for convenience.[164][166] Most protists are unicellular, which are also known as microbial eukaryotes.[164]
The alveolates are mostly photosynthetic unicellular protists that possess sacs called alveoli (hence their name alveolates) that are located beneath their cell membrane, providing support for the cell surface.[164] Alveolates comprise several groups such as dinoflagellates, apicomplexans, and ciliates. Dinoflagellates are photosynthetic and can be found in the ocean where they play a role as primary producers of organic matter.[164] Apicomplexans are parasitic alveolates that possess an apical complex, which is a group of organelles located in the apical end of the cell.[164] This complex allows apicomplexans to invade their hosts' tissues. Ciliates are alveolates that possess numerous hair-like structure called cilia. A defining characteristic of ciliates is the presence of two types of nuclei in each ciliate cell. A commonly studied ciliate is the paramecium.[164]
The excavates are groups of protists that began to diversify approximately 1.5 billion years ago shortly after the origin of the eukaryotes.[164] Some excavates do not possess mitochondria, which are thought to have been lost over the course of evolution as these protists still possess nuclear genes that are associated with mitochondria.[164] The excavates comprise several groups such as diplomonads, parabasalids, heteroloboseans, euglenids, and kinetoplastids.[164]
Stramenopiles, most of which can be characterized by the presence of tubular hairs on the longer of their two flagella, include diatoms and brown algae.[164] Diatoms are primary producers and contribute about one-fifth of all photosynthetic carbon fixation, making them a major component of phytoplankton.[164]
Rhizarians are mostly unicellular and aquatic protists that typically contain long, thin pseudopods.[164] The rhizarians comprise three main groups: cercozoans, foraminiferans, and radiolarians.[164]
Amoebozoans are protists with a body form characterized by the presence lobe-shaped pseudopods, which help them to move.[164] They include groups such as loboseans and slime molds (e.g., plasmodial slime mold and cellular slime molds).[164]
Plant diversity
_version_2.png.webp)
Plants are mainly multicellular organisms, predominantly photosynthetic eukaryotes of the kingdom Plantae, which would exclude fungi and some algae. A shared derived trait (or synapomorphy) of Plantae is the primary endosymbiosis of a cyanobacterium into an early eukaryote about one billion years ago, which gave rise to chloroplasts.[167] The first several clades that emerged following primary endosymbiosis were aquatic and most of the aquatic photosynthetic eukaryotic organisms are collectively described as algae, which is a term of convenience as not all algae are closely related.[167] Algae comprise several distinct clades such as glaucophytes, which are microscopic freshwater algae that may have resembled in form to the early unicellular ancestor of Plantae.[167] Unlike glaucophytes, the other algal clades such as red and green algae are multicellular. Green algae comprise three major clades: chlorophytes, coleochaetophytes, and stoneworts.[167]
Land plants (embryophytes) first appeared in terrestrial environments approximately 450 to 500 million years ago.[167] A synapomorphy of land plants is an embryo that develops under the protection of tissues of its parent plant.[167] Land plants comprise ten major clades, seven of which constitute a single clade known as vascular plants (or tracheophytes) as they all have tracheids, which are fluid-conducting cells, and a well-developed system that transports materials throughout their bodies.[167] In contrast, the other three clades are nonvascular plants as they do not have tracheids.[167] They also do not constitute a single clade.[167]
Nonvascular plants include liverworts, mosses, and hornworts. They tend to be found in areas where water is readily available.[167] Most live on soil or even on vascular plants themselves. Some can grow on bare rock, tree trunks that are dead or have fallen, and even buildings.[167] Most nonvascular plants are terrestrial, with a few living in freshwater environments and none living in the oceans.[167]
The seven clades (or divisions) that make up vascular plants include horsetails and ferns, which together can be grouped as a single clade called monilophytes.[167] Seed plants (or spermatophyte) comprise the other five divisions, four of which are grouped as gymnosperms and one is angiosperms. Gymnosperms includes conifers, cycads, Ginkgo, and gnetophytes. Gymnosperm seeds develop either on the surface of scales or leaves, which are often modified to form cones, or solitary as in yew, Torreya, Ginkgo.[168] Angiosperms are the most diverse group of land plants, with 64 orders, 416 families, approximately 13,000 known genera and 300,000 known species.[169] Like gymnosperms, angiosperms are seed-producing plants. They are distinguished from gymnosperms by having characteristics such as flowers, endosperm within their seeds, and production of fruits that contain the seeds.
Fungi

Fungi are eukaryotic organisms that digest foods outside of their bodies.[170] They do so through a process called absorptive heterotrophy whereby they would first secrete digestive enzymes that break down large food molecules before absorbing them through their cell membranes. Many fungi are also saprobes as they are able to take in nutrients from dead organic matter and are hence, the principal decomposers in ecological systems.[170] Some fungi are parasites by absorbing nutrients from living hosts while others are mutualists.[170] Fungi, along with two other lineages, choanoflagellates and animals, can be grouped as opisthokonts. A synapomorphy that distinguishes fungi from other two opisthokonts is the presence of chitin in their cell walls.[170]
Most fungi are multicellular but some are unicellular such as yeasts, which live in liquid or moist environments and are able to absorb nutrients directly into their cell surfaces.[170] Multicellular fungi, on the other hand, have a body called mycelium, which is composed of a mass of individual tubular filaments called hyphae that allows for nutrient absorption to occur.[170]
Fungi can be divided into six major groups based on their life cycles: microsporidia, chytrids, zygospore fungi (Zygomycota), arbuscular mycorrhizal fungi (Glomeromycota), sac fungi (Ascomycota), and club fungi (Basidiomycota).[170] Fungi are classified by the particular processes of sexual reproduction they use. The usual cellular products of meiosis during sexual reproduction are spores that are adapted to survive inclement times and to spread. A principal adaptive benefit of meiosis during sexual reproduction in the Ascomycota and Basidiomycota was proposed to be the repair of DNA damage through meiotic recombination.[171]
The fungus kingdom encompasses an enormous diversity of taxa with varied ecologies, life cycle strategies, and morphologies ranging from unicellular aquatic chytrids to large mushrooms. However, little is known of the true biodiversity of Kingdom Fungi, which has been estimated at 2.2 million to 3.8 million species.[172] Of these, only about 148,000 have been described,[173] with over 8,000 species known to be detrimental to plants and at least 300 that can be pathogenic to humans.[174]
Animal diversity

Animals are multicellular eukaryotic organisms that form the kingdom Animalia. With few exceptions, animals consume organic material, breathe oxygen, are able to move, can reproduce sexually, and grow from a hollow sphere of cells, the blastula, during embryonic development. Over 1.5 million living animal species have been described—of which around 1 million are insects—but it has been estimated there are over 7 million animal species in total. They have complex interactions with each other and their environments, forming intricate food webs.
Animals can be distinguished into two groups based on their developmental characteristics.[175] For instance, embryos of diploblastic animals such as ctenophores, placeozoans, and cnidarians have two cell layers (ectoderm and endoderm) whereas the embryos of triploblastic animals have three tissue layers (ectoderm, mesoderm, and endoderm), which is a synapomorphy of these animals.[175] Triploblastic animals can be further divided into two major clades based on based on the pattern of gastrulation, whereby a cavity called a blastopore is formed from the indentation of a blastula. In protostomes, the blastopore gives rise to the mouth, which is then followed by the formation of the anus.[175] In deuterostomes, the blastopore gives rise to the anus, followed by the formation of the mouth.[175]
Animals can also be differentiated based on their body plan, specifically with respect to four key features: symmetry, body cavity, segmentation, and appendages.[175] The bodies of most animals are symmetrical, with symmetry being either radial or bilateral.[175] Triploblastic animals can be divided into three types based on their body cavity: acoelomate, pseudocoelomate, and coelomate.[175] Segmentation can be observed in the bodies of many animals, which allows for specialization of different parts of the body as well as allowing the animal to change the shape of its body to control its movements.[175] Finally, animals can be distinguished based on the type and location of their appendages such as antennae for sensing the environment or claws for capturing prey.[175]
Sponges, the members of the phylum Porifera, are a basal Metazoa (animal) clade as a sister of the diploblasts.[176][177][178][179][180] They are multicellular organisms that have bodies full of pores and channels allowing water to circulate through them, consisting of jelly-like mesohyl sandwiched between two thin layers of cells.
The majority (~97%) of animal species are invertebrates,[181] which are animals that do not have a vertebral column (or backbone or spine), derived from the notochord. This includes all animals apart from the subphylum Vertebrata. Familiar examples of invertebrates include sponges, cnidarians (hydras, jellyfishes, sea anemones, and corals), mollusks (chitons, snail, bivalves, squids, and octopuses), annelids (earthworms and leeches), and arthropods (insects, arachnids, crustaceans, and myriapods). Many invertebrate taxa have a greater number and variety of species than the entire subphylum of Vertebrata.[182]
In contrast, vertebrates comprise all species of animals within the subphylum Vertebrata, which are chordates with vertebral columns. These animals have four key features, which are an anterior skull with a brain, a rigid internal skeleton supported by a vertebral column that encloses a spinal cord, internal organs suspended in a coelom, and a well-developed circulatory system driven by a single large heart.[175] Vertebrates represent the overwhelming majority of the phylum Chordata, with currently about 69,963 species described.[183] Vertebrates comprise different major groups that include jawless fishes (not including hagfishes), jawed vertebrates such as cartilaginous fishes (sharks, rays, and ratfish), bony fishes, tetrapods such as amphibians, reptiles, birds, and mammals.[175]
The two remaining groups of jawless fishes that have survived beyond the Devonian period are hagfishes and lamprey, which are collectively known as cyclostomes (for circled mouths).[175] Both groups of animals have elongated eel-like bodies with no paired fins.[175] However, because hagfishes have a weak circulatory system with three accessory hearts, a partial skull with no cerebellum, no jaws or stomach, and no jointed vertebrae, some biologists do not classify them as vertebrates but instead as a sister group of vertebrates.[175] In contrast, lampreys have a complete skull and a distinct vertebrae that is cartilaginous.[175]
Mammals have four key features that distinguish them from other animals such as sweat glands, mammary glands, hair, and a four-chambered heart.[175] Small and medium-sized mammals used to co-exist with large dinosaurs in much of the Mesozoic era but soon radiated following the mass extinction of dinosaurs at the end of the Cretaceous period.[175] There are approximately 57,000 mammal species, which can be divided into two primary groups: prototherians and therians. Prototherians do not possess nipples on their mammary but instead secrete milk onto their skin, allowing their offspring to lap if off their furs.[175] They also lack a placenta, lays eggs, and have sprawling legs. Currently, there only five known species of prototherians (platypus and four species of echidnas).[175] The therian clade is viviparous and can be further divided into two groups: marsupials and eutherians.[175] Marsupial females have a ventral pouch to carry and feed their offspring. Eutherians form the majority of mammals and include major groups such as rodents, bats, even-toed ungulates and cetaceans, shrews and moles, primates, carnivores, rabbits, African insectivores, spiny insectivores, armadillos, treeshrews, odd-toed ungulates, long-nosed insectivores, anteaters and sloths, pangolins, hyraxes, sirenians, elephants, colugos, and aardvark.[175]
A split in the primate lineage occurred approximately 90 million years ago during the Cretaceous, which brought about two major clades: prosimians and anthropoids.[175] The prosimians include lemurs, lorises, and galagos whereas the anthropoids comprise tarsiers, New World monkeys, Old World monkeys, and apes.[175] Apes separated from Old World monkeys about 35 million years ago, with various species living in Africa, Europe, and Asia between 22 and 5.5 million years ago.[175] The modern descendants of these animals include chimpanzees and gorillas in Africa, gibbons and orangutans in Asia, and humans worldwide. A split in the ape lineage occurred about six million years ago in Africa, which resulted in the emergence of chimpanzees as one group and a hominid clade as another group that includes humans and their extinct relatives.[175] Bipedalism emerged in the earliest protohominids known as ardipithecines. As an adaptation, bipedalism conferred three advantages. First, it enabled the ardipithecines to use their forelimbs to manipulate and carry objects while working.[175] Second, it elevated the animal's eyes to spot preys or predators over tall vegetation.[175] Finally, bipedalism is more energetically efficient than quadrupedal locomotion.[175]
Viruses

Viruses are submicroscopic infectious agents that replicate inside the cells of organisms.[184] Viruses infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.[185][186] More than 6,000 virus species have been described in detail.[187] Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity.[188][189]
When infected, a host cell is forced to rapidly produce thousands of identical copies of the original virus. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent particles, or virions, consisting of the genetic material (DNA or RNA), a protein coat called capsid, and in some cases an outside envelope of lipids. The shapes of these virus particles range from simple helical and icosahedral forms to more complex structures. Most virus species have virions too small to be seen with an optical microscope, as they are one-hundredth the size of most bacteria.
The origins of viruses in the evolutionary history of life are unclear: some may have evolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria. In evolution, viruses are an important means of horizontal gene transfer, which increases genetic diversity in a way analogous to sexual reproduction.[190] Because viruses possess some but not all characteristics of life, they have been described as "organisms at the edge of life",[191] and as self-replicators.[192]
Viruses can spread in many ways. One transmission pathway is through disease-bearing organisms known as vectors: for example, viruses are often transmitted from plant to plant by insects that feed on plant sap, such as aphids; and viruses in animals can be carried by blood-sucking insects. Influenza viruses are spread by coughing and sneezing. Norovirus and rotavirus, common causes of viral gastroenteritis, are transmitted by the faecal–oral route, passed by hand-to-mouth contact or in food or water. Viral infections in animals provoke an immune response that usually eliminates the infecting virus. Immune responses can also be produced by vaccines, which confer an artificially acquired immunity to the specific viral infection.
Plant form and function
Plant body

The plant body is made up of organs that can be organized into two major organ systems: a root system and a shoot system.[193] The root system anchors the plants into place. The roots themselves absorb water and minerals and store photosynthetic products. The shoot system is composed of stem, leaves, and flowers. The stems hold and orient the leaves to the sun, which allow the leaves to conduct photosynthesis. The flowers are shoots that have been modified for reproduction. Shoots are composed of phytomers, which are functional units that consist of a node carrying one or more leaves, internode, and one or more buds.
A plant body has two basic patterns (apical–basal and radial axes) that been established during embryogenesis.[193] Cells and tissues are arranged along the apical-basal axis from root to shoot whereas the three tissue systems (dermal, ground, and vascular) that make up a plant's body are arranged concentrically around its radial axis.[193] The dermal tissue system forms the epidermis (or outer covering) of a plant, which is usually a single cell layer that consists of cells that have differentiated into three specialized structures: stomata for gas exchange in leaves, trichomes (or leaf hair) for protection against insects and solar radiation, and root hairs for increased surface areas and absorption of water and nutrients. The ground tissue makes up virtually all the tissue that lies between the dermal and vascular tissues in the shoots and roots. It consists of three cell types: Parenchyma, collenchyma, and sclerenchyma cells. Finally, the vascular tissues are made up of two constituent tissues: xylem and phloem. The xylem is made up of two conducting cells called tracheids and vessel elements whereas the phloem is characterized by the presence of sieve tube elements and companion cells.[193]
Plant nutrition and transport

Like all other organisms, plants are primarily made up of water and other molecules containing elements that are essential to life.[194] The absence of specific nutrients (or essential elements), many of which have been identified in hydroponic experiments, can disrupt plant growth and reproduction. The majority of plants are able to obtain these nutrients from solutions that surrounds their roots in the soil.[194] Continuous leaching and harvesting of crops can deplete the soil of its nutrients, which can be restored with the use of fertilizers. Carnivorous plants such as Venus flytraps are able to obtain nutrients by digesting other arthropods whereas parasitic plants such as mistletoes can parasitize other plants for water and nutrients.
Plants need water to conduct photosynthesis, transport solutes between organs, cool their leaves by evaporation, and maintain internal pressures that support their bodies.[194] Water is able to diffuse in and out of plant cells by osmosis. The direction of water movement across a semipermeable membrane is determined by the water potential across that membrane.[194] Water is able to diffuse across a root cell's membrane through aquaporins whereas solutes are transported across by the membrane by ion channels and pumps. In vascular plants, water and solutes are able to enter the xylem, a vascular tissue, by way of an apoplast and symplast. Once in the xylem, the water and minerals are distributed upward by transpiration from the soil to the aerial parts of the plant.[167][194] In contrast, the phloem, another vascular tissue, distributes carbohydrates (e.g., sucrose) and other solutes such as hormones by translocation from a source (e.g., mature leaf or root) in which they were produced to a sink (e.g., root, flower, or developing fruit) in which they will be used and stored.[194] Sources and sinks can switch roles, depending on the amount of carbohydrates accumulated or mobilized for the nourishment of other organs.
Plant development
Plant development is regulated by environmental cues and the plant's own receptors, hormones, and genome.[195] Morever, they have several characteristics that allow them to obtain resources for growth and reproduction such as meristems, post-embryonic organ formation, and differential growth.
Development begins with a seed, which is an embryonic plant enclosed in a protective outer covering. Most plant seeds are usually dormant, a condition in which the seed's normal activity is suspended.[195] Seed dormancy may last may last weeks, months, years, and even centuries. Dormancy is broken once conditions are favorable for growth, and the seed will begin to sprout, a process called germination. Imbibition is the first step in germination, whereby water is absorbed by the seed. Once water is absorbed, the seed undergoes metabolic changes whereby enzymes are activated and RNA and proteins are synthesized. Once the seed germinates, it obtains carbohydrates, amino acids, and small lipids that serve as building blocks for its development. These monomers are obtained from the hydrolysis of starch, proteins, and lipids that are stored in either the cotyledons or endosperm. Germination is completed once embryonic roots called radicle have emerged from the seed coat. At this point, the developing plant is called a seedling and its growth is regulated by its own photoreceptor proteins and hormones.[195]
Unlike animals in which growth is determinate, i.e., ceases when the adult state is reached, plant growth is indeterminate as it is an open-ended process that could potentially be lifelong.[193] Plants grow in two ways: primary and secondary. In primary growth, the shoots and roots are formed and lengthened. The apical meristem produces the primary plant body, which can be found in all seed plants. During secondary growth, the thickness of the plant increases as the lateral meristem produces the secondary plant body, which can be found in woody eudicots such as trees and shrubs. Monocots do not go through secondary growth.[193] The plant body is generated by a hierarchy of meristems. The apical meristems in the root and shoot systems give rise to primary meristems (protoderm, ground meristem, and procambium), which in turn, give rise to the three tissue systems (dermal, ground, and vascular).
Plant reproduction

Most angiosperms (or flowering plants) engage in sexual reproduction.[196] Their flowers are organs that facilitate reproduction, usually by providing a mechanism for the union of sperm with eggs. Flowers may facilitate two types of pollination: self-pollination and cross-pollination. Self-pollination occurs when the pollen from the anther is deposited on the stigma of the same flower, or another flower on the same plant. Cross-pollination is the transfer of pollen from the anther of one flower to the stigma of another flower on a different individual of the same species. Self-pollination happened in flowers where the stamen and carpel mature at the same time, and are positioned so that the pollen can land on the flower's stigma. This pollination does not require an investment from the plant to provide nectar and pollen as food for pollinators.[197]
Plant responses
Like animals, plants produce hormones in one part of its body to signal cells in another part to respond. The ripening of fruit and loss of leaves in the winter are controlled in part by the production of the gas ethylene by the plant. Stress from water loss, changes in air chemistry, or crowding by other plants can lead to changes in the way a plant functions. These changes may be affected by genetic, chemical, and physical factors.
To function and survive, plants produce a wide array of chemical compounds not found in other organisms. Because they cannot move, plants must also defend themselves chemically from herbivores, pathogens and competition from other plants. They do this by producing toxins and foul-tasting or smelling chemicals. Other compounds defend plants against disease, permit survival during drought, and prepare plants for dormancy, while other compounds are used to attract pollinators or herbivores to spread ripe seeds.
Many plant organs contain different types of photoreceptor proteins, each of which reacts very specifically to certain wavelengths of light.[198] The photoreceptor proteins relay information such as whether it is day or night, duration of the day, intensity of light available, and the source of light. Shoots generally grow towards light, while roots grow away from it, responses known as phototropism and skototropism, respectively. They are brought about by light-sensitive pigments like phototropins and phytochromes and the plant hormone auxin.[199] Many flowering plants bloom at the appropriate time because of light-sensitive compounds that respond to the length of the night, a phenomenon known as photoperiodism.
In addition to light, plants can respond to other types of stimuli. For instance, plants can sense the direction of gravity to orient themselves correctly. They can respond to mechanical stimulation.[200]
Animal form and function
General features

The cells in each animal body are bathed in interstitial fluid, which make up the cell's environment. This fluid and all its characteristics (e.g., temperature, ionic composition) can be described as the animal's internal environment, which is in contrast to the external environment that encompasses the animal's outside world.[201] Animals can be classified as either regulators or conformers. Animals such as mammals and birds are regulators as they are able to maintain a constant internal environment such as body temperature despite their environments changing. These animals are also described as homeotherms as they exhibit thermoregulation by keeping their internal body temperature constant. In contrast, animals such as fishes and frogs are conformers as they adapt their internal environment (e.g., body temperature) to match their external environments. These animals are also described as poikilotherms or ectotherms as they allow their body temperatures to match their external environments. In terms of energy, regulation is more costly than conformity as an animal expands more energy to maintain a constant internal environment such as increasing its basal metabolic rate, which is the rate of energy consumption.[201] Similarly, homeothermy is more costly than poikilothermy. Homeostasis is the stability of an animal's internal environment, which is maintained by negative feedback loops.[201][202]
The body size of terrestrial animals vary across different species but their use of energy does not scale linearly according to their size.[201] Mice, for example, are able to consume three times more food than rabbits in proportion to their weights as the basal metabolic rate per unit weight in mice is greater than in rabbits.[201] Physical activity can also increase an animal's metabolic rate. When an animal runs, its metabolic rate increases linearly with speed.[201] However, the relationship is non-linear in animals that swim or fly. When a fish swims faster, it encounters greater water resistance and so its metabolic rates increases exponential.[201] Alternatively, the relationship of flight speeds and metabolic rates is U-shaped in birds.[201] At low flight speeds, a bird must maintain a high metabolic rates to remain airborne. As it speeds up its flight, its metabolic rate decreases with the aid of air rapidly flows over its wings. However, as it increases in its speed even further, its high metabolic rates rises again due to the increased effort associated with rapid flight speeds. Basal metabolic rates can be measured based on an animal's rate of heat production.
Water and salt balance

An animal's body fluids have three properties: osmotic pressure, ionic composition, and volume.[203] Osmotic pressures determine the direction of the diffusion of water (or osmosis), which moves from a region where osmotic pressure (total solute concentration) is low to a region where osmotic pressure (total solute concentration) is high. Aquatic animals are diverse with respect to their body fluid compositions and their environments. For example, most invertebrate animals in the ocean have body fluids that are isosmotic with seawater. In contrast, ocean bony fishes have body fluids that are hyposmotic to seawater. Finally, freshwater animals have body fluids that are hyperosmotic to fresh water. Typical ions that can be found in an animal's body fluids are sodium, potassium, calcium, and chloride. The volume of body fluids can be regulated by excretion. Vertebrate animals have kidneys, which are excretory organs made up of tiny tubular structures called nephrons, which make urine from blood plasma. The kidneys' primary function is to regulate the composition and volume of blood plasma by selectively removing material from the blood plasma itself. The ability of xeric animals such as kangaroo rats to minimize water loss by producing urine that is 10–20 times concentrated than their blood plasma allows them to adapt in desert environments that receive very little precipitation.[203]
Nutrition and digestion

Animals are heterotrophs as they feed on other organisms to obtain energy and organic compounds.[204] They are able to obtain food in three major ways such as targeting visible food objects, collecting tiny food particles, or depending on microbes for critical food needs. The amount of energy stored in food can be quantified based on the amount of heat (measured in calories or kilojoules) emitted when the food is burnt in the presence of oxygen. If an animal were to consume food that contains an excess amount of chemical energy, it will store most of that energy in the form of lipids for future use and some of that energy as glycogen for more immediate use (e.g., meeting the brain's energy needs).[204] The molecules in food are chemical building blocks that are needed for growth and development. These molecules include nutrients such as carbohydrates, fats, and proteins. Vitamins and minerals (e.g., calcium, magnesium, sodium, and phosphorus) are also essential. The digestive system, which typically consist of a tubular tract that extends from the mouth to the anus, is involved in the breakdown (or digestion) of food into small molecules as it travels down peristaltically through the gut lumen shortly after it has been ingested. These small food molecules are then absorbed into the blood from the lumen, where they are then distributed to the rest of the body as building blocks (e.g., amino acids) or sources of energy (e.g., glucose).[204]
In addition to their digestive tracts, vertebrate animals have accessory glands such as a liver and pancreas as part of their digestive systems.[204] The processing of food in these animals begins in the foregut, which includes the mouth, esophagus, and stomach. Mechanical digestion of food starts in the mouth with the esophagus serving as a passageway for food to reach the stomach, where it is stored and disintegrated (by the stomach's acid) for further processing. Upon leaving the stomach, food enters into the midgut, which is the first part of the intestine (or small intestine in mammals) and is the principal site of digestion and absorption. Food that does not get absorbed are stored as indigestible waste (or feces) in the hindgut, which is the second part of the intestine (or large intestine in mammals). The hindgut then completes the reabsorption of needed water and salt prior to eliminating the feces from the rectum.[204]
Breathing

The respiratory system consists of specific organs and structures used for gas exchange in animals. The anatomy and physiology that make this happen varies greatly, depending on the size of the organism, the environment in which it lives and its evolutionary history. In land animals the respiratory surface is internalized as linings of the lungs.[205] Gas exchange in the lungs occurs in millions of small air sacs; in mammals and reptiles these are called alveoli, and in birds they are known as atria. These microscopic air sacs have a very rich blood supply, thus bringing the air into close contact with the blood.[206] These air sacs communicate with the external environment via a system of airways, or hollow tubes, of which the largest is the trachea, which branches in the middle of the chest into the two main bronchi. These enter the lungs where they branch into progressively narrower secondary and tertiary bronchi that branch into numerous smaller tubes, the bronchioles. In birds the bronchioles are termed parabronchi. It is the bronchioles, or parabronchi that generally open into the microscopic alveoli in mammals and atria in birds. Air has to be pumped from the environment into the alveoli or atria by the process of breathing, which involves the muscles of respiration.
Circulation

A circulatory system usually consists of a muscular pump such as a heart, a fluid (blood), and system of blood vessels that deliver it.[207][208] Its principal function is to transport blood and other substances to and from cells and tissues. There are two types of circulatory systems: open and closed. In open circulatory systems, blood exits blood vessels as it circulates throughout the body whereas in closed circulatory system, blood is contained within the blood vessels as it circulates. Open circulatory systems can be observed in invertebrate animals such as arthropods (e.g., insects, spiders, and lobsters) whereas closed circulatory systems can be found in vertebrate animals such as fishes, amphibians, and mammals. Circulation in animals occur between two types of tissues: systemic tissues and breathing (or pulmonary) organs.[207] Systemic tissues are all the tissues and organs that make up an animal's body other than its breathing organs. Systemic tissues take up oxygen but adds carbon dioxide to the blood whereas a breathing organs takes up carbon dioxide but add oxygen to the blood.[209] In birds and mammals, the systemic and pulmonary systems are connected in series.
In the circulatory system, blood is important because it is the means by which oxygen, carbon dioxide, nutrients, hormones, agents of immune system, heat, wastes, and other commodities are transported.[207] In annelids such as earthworms and leeches, blood is propelled by peristaltic waves of contractions of the heart muscles that make up the blood vessels. Other animals such as crustaceans (e.g., crayfish and lobsters), have more than one heart to propel blood throughout their bodies. Vertebrate hearts are multichambered and are able to pump blood when their ventricles contract at each cardiac cycle, which propels blood through the blood vessels.[207] Although vertebrate hearts are myogenic, their rate of contraction (or heart rate) can be modulated by neural input from the body's autonomic nervous system.
Muscle and movement

In vertebrates, the muscular system consists of skeletal, smooth and cardiac muscles. It permits movement of the body, maintains posture and circulates blood throughout the body.[210] Together with the skeletal system, it forms the musculoskeletal system, which is responsible for the movement of vertebrate animals.[211] Skeletal muscle contractions are neurogenic as they require synaptic input from motor neurons. A single motor neuron is able to innervate multiple muscle fibers, thereby causing the fibers to contract at the same time. Once innervated, the protein filaments within each skeletal muscle fiber slide past each other to produce a contraction, which is explained by the sliding filament theory. The contraction produced can be described as a twitch, summation, or tetanus, depending on the frequency of action potentials. Unlike skeletal muscles, contractions of smooth and cardiac muscles are myogenic as they are initiated by the smooth or heart muscle cells themselves instead of a motor neuron. Nevertheless, the strength of their contractions can be modulated by input from the autonomic nervous system. The mechanisms of contraction are similar in all three muscle tissues.
In invertebrates such as earthworms and leeches, circular and longitudinal muscles cells form the body wall of these animals and are responsible for their movement.[212] In an earthworm that is moving through a soil, for example, contractions of circular and longitudinal muscles occur reciprocally while the coelomic fluid serves as a hydroskeleton by maintaining turgidity of the earthworm.[213] Other animals such as mollusks, and nematodes, possess obliquely striated muscles, which contain bands of thick and thin filaments that are arranged helically rather than transversely, like in vertebrate skeletal or cardiac muscles.[214] Advanced insects such as wasps, flies, bees, and beetles possess asynchronous muscles that constitute the flight muscles in these animals.[214] These flight muscles are often called fibrillar muscles because they contain myofibrils that are thick and conspicuous.[215]
Nervous system
Most multicellular animals have nervous systems[217] that allow them to sense from and respond to their environments. A nervous system is a network of cells that processes sensory information and generates behaviors. At the cellular level, the nervous system is defined by the presence of neurons, which are cells specialized to handle information.[218] They can transmit or receive information at sites of contacts called synapses.[218] More specifically, neurons can conduct nerve impulses (or action potentials) that travel along their thin fibers called axons, which can then be transmitted directly to a neighboring cell through electrical synapses or cause chemicals called neurotransmitters to be released at chemical synapses. According to the sodium theory, these action potentials can be generated by the increased permeability of the neuron's cell membrane to sodium ions.[219] Cells such as neurons or muscle cells may be excited or inhibited upon receiving a signal from another neuron. The connections between neurons can form neural pathways, neural circuits, and larger networks that generate an organism's perception of the world and determine its behavior. Along with neurons, the nervous system contains other specialized cells called glia or glial cells, which provide structural and metabolic support.
In vertebrates, the nervous system comprises the central nervous system (CNS), which includes the brain and spinal cord, and the peripheral nervous system (PNS), which consists of nerves that connect the CNS to every other part of the body. Nerves that transmit signals from the CNS are called motor nerves or efferent nerves, while those nerves that transmit information from the body to the CNS are called sensory nerves or afferent nerves. Spinal nerves are mixed nerves that serve both functions. The PNS is divided into three separate subsystems, the somatic, autonomic, and enteric nervous systems. Somatic nerves mediate voluntary movement. The autonomic nervous system is further subdivided into the sympathetic and the parasympathetic nervous systems. The sympathetic nervous system is activated in cases of emergencies to mobilize energy, while the parasympathetic nervous system is activated when organisms are in a relaxed state. The enteric nervous system functions to control the gastrointestinal system. Both autonomic and enteric nervous systems function involuntarily. Nerves that exit directly from the brain are called cranial nerves while those exiting from the spinal cord are called spinal nerves.
Many animals have sense organs that can detect their environment. These sense organs contain sensory receptors, which are sensory neurons that convert stimuli into electrical signals.[220] Mechanoreceptors, for example, which can be found in skin, muscle, and hearing organs, generate action potentials in response to changes in pressures.[220][221] Photoreceptor cells such as rods and cones, which are part of the vertebrate retina, can respond to specific wavelengths of light.[220][221] Chemoreceptors detect chemicals in the mouth (taste) or in the air (smell).[221]
Hormonal control
Hormones are signaling molecules transported in the blood to distant organs to regulate their function.[222][223] Hormones are secreted by internal glands that are part of an animal's endocrine system. In vertebrates, the hypothalamus is the neural control center for all endocrine systems. In humans specifically, the major endocrine glands are the thyroid gland and the adrenal glands. Many other organs that are part of other body systems have secondary endocrine functions, including bone, kidneys, liver, heart and gonads. For example, kidneys secrete the endocrine hormone erythropoietin. Hormones can be amino acid complexes, steroids, eicosanoids, leukotrienes, or prostaglandins.[224] The endocrine system can be contrasted to both exocrine glands, which secrete hormones to the outside of the body, and paracrine signaling between cells over a relatively short distance. Endocrine glands have no ducts, are vascular, and commonly have intracellular vacuoles or granules that store their hormones. In contrast, exocrine glands, such as salivary glands, sweat glands, and glands within the gastrointestinal tract, tend to be much less vascular and have ducts or a hollow lumen.
Animal reproduction

Animals can reproduce in one of two ways: asexual and sexual. Nearly all animals engage in some form of sexual reproduction.[225] They produce haploid gametes by meiosis. The smaller, motile gametes are spermatozoa and the larger, non-motile gametes are ova.[226] These fuse to form zygotes,[227] which develop via mitosis into a hollow sphere, called a blastula. In sponges, blastula larvae swim to a new location, attach to the seabed, and develop into a new sponge.[228] In most other groups, the blastula undergoes more complicated rearrangement.[229] It first invaginates to form a gastrula with a digestive chamber and two separate germ layers, an external ectoderm and an internal endoderm.[230] In most cases, a third germ layer, the mesoderm, also develops between them.[231] These germ layers then differentiate to form tissues and organs.[232] Some animals are capable of asexual reproduction, which often results in a genetic clone of the parent. This may take place through fragmentation; budding, such as in Hydra and other cnidarians; or parthenogenesis, where fertile eggs are produced without mating, such as in aphids.[233][234]
Animal development
Animal development begins with the formation of a zygote that results from the fusion of a sperm and egg during fertilization.[235] The zygote undergoes a rapid multiple rounds of mitotic cell period of cell divisions called cleavage, which forms a ball of similar cells called a blastula. Gastrulation occurs, whereby morphogenetic movements convert the cell mass into a three germ layers that comprise the ectoderm, mesoderm and endoderm.
The end of gastrulation signals the beginning of organogenesis, whereby the three germ layers form the internal organs of the organism.[236] The cells of each of the three germ layers undergo differentiation, a process where less-specialized cells become more-specialized through the expression of a specific set of genes. Cellular differentiation is influenced by extracellular signals such as growth factors that are exchanged to adjacent cells, which is called juxtracrine signaling, or to neighboring cells over short distances, which is called paracrine signaling.[237][238] Intracellular signals consist of a cell signaling itself (autocrine signaling), also play a role in organ formation. These signaling pathways allows for cell rearrangement and ensures that organs form at specific sites within the organism.[236][239]
Immune system

The immune system is a network of biological processes that detects and responds to a wide variety of pathogens. Many species have two major subsystems of the immune system. The innate immune system provides a preconfigured response to broad groups of situations and stimuli. The adaptive immune system provides a tailored response to each stimulus by learning to recognize molecules it has previously encountered. Both use molecules and cells to perform their functions.
Nearly all organisms have some kind of immune system. Bacteria have a rudimentary immune system in the form of enzymes that protect against virus infections. Other basic immune mechanisms evolved in ancient plants and animals and remain in their modern descendants. These mechanisms include phagocytosis, antimicrobial peptides called defensins, and the complement system. Jawed vertebrates, including humans, have even more sophisticated defense mechanisms, including the ability to adapt to recognize pathogens more efficiently. Adaptive (or acquired) immunity creates an immunological memory leading to an enhanced response to subsequent encounters with that same pathogen. This process of acquired immunity is the basis of vaccination.
Animal behavior

Behaviors play a central a role in animals' interaction with each other and with their environment.[240] They are able to use their muscles to approach one another, vocalize, seek shelter, and migrate. An animal's nervous system activates and coordinates its behaviors. Fixed action patterns, for instance, are genetically determined and stereotyped behaviors that occur without learning.[240][241] These behaviors are under the control of the nervous system and can be quite elaborate.[240] Examples include the pecking of kelp gull chicks at the red dot on their mother's beak. Other behaviors that have emerged as a result of natural selection include foraging, mating, and altruism.[242] In addition to evolved behavior, animals have evolved the ability to learn by modifying their behaviors as a result of early individual experiences.[240]
Ecology
Ecology is the study of the distribution and abundance of life, the interaction between organisms and their environment.[243]
Ecosystems

The community of living (biotic) organisms in conjunction with the nonliving (abiotic) components (e.g., water, light, radiation, temperature, humidity, atmosphere, acidity, and soil) of their environment is called an ecosystem.[244][245][246] These biotic and abiotic components are linked together through nutrient cycles and energy flows.[247] Energy from the sun enters the system through photosynthesis and is incorporated into plant tissue. By feeding on plants and on one another, animals play an important role in the movement of matter and energy through the system. They also influence the quantity of plant and microbial biomass present. By breaking down dead organic matter, decomposers release carbon back to the atmosphere and facilitate nutrient cycling by converting nutrients stored in dead biomass back to a form that can be readily used by plants and other microbes.[248]
The Earth's physical environment is shaped by solar energy and topography.[246] The amount of solar energy input varies in space and time due to the spherical shape of the Earth and its axial tilt. Variation in solar energy input drives weather and climate patterns. Weather is the day-to-day temperature and precipitation activity, whereas climate is the long-term average of weather, typically averaged over a period of 30 years.[249][250] Variation in topography also produces environmental heterogeneity. On the windward side of a mountain, for example, air rises and cools, with water changing from gaseous to liquid or solid form, resulting in precipitation such as rain or snow.[246] As a result, wet environments allow for lush vegetation to grow. In contrast, conditions tend to be dry on the leeward side of a mountain due to the lack of precipitation as air descends and warms, and moisture remains as water vapor in the atmosphere. Temperature and precipitation are the main factors that shape terrestrial biomes.
Populations

A population is the number of organisms of the same species that occupy an area and reproduce from generation to generation.[251][252][253][254][255] Its abundance can be measured using population density, which is the number of individuals per unit area (e.g., land or tree) or volume (e.g., sea or air).[251] Given that it is usually impractical to count every individual within a large population to determine its size, population size can be estimated by multiplying population density by the area or volume. Population growth during short-term intervals can be determined using the population growth rate equation, which takes into consideration birth, death, and immigration rates. In the longer term, the exponential growth of a population tends to slow down as it reaches its carrying capacity, which can be modeled using the logistic equation.[252] The carrying capacity of an environment is the maximum population size of a species that can be sustained by that specific environment, given the food, habitat, water, and other resources that are available.[256] The carrying capacity of a population can be affected by changing environmental conditions such as changes in the availability resources and the cost of maintaining them. In human populations, new technologies such as the Green revolution have helped increase the Earth's carrying capacity for humans over time, which has stymied the attempted predictions of impending population decline, the famous of which was by Thomas Malthus in the 18th century.[251]
Communities

A community is a group of populations of two or more different species occupying the same geographical area at the same time. A biological interaction is the effect that a pair of organisms living together in a community have on each other. They can be either of the same species (intraspecific interactions), or of different species (interspecific interactions). These effects may be short-term, like pollination and predation, or long-term; both often strongly influence the evolution of the species involved. A long-term interaction is called a symbiosis. Symbioses range from mutualism, beneficial to both partners, to competition, harmful to both partners.[258]
Every species participates as a consumer, resource, or both in consumer–resource interactions, which form the core of food chains or food webs.[259] There are different trophic levels within any food web, with the lowest level being the primary producers (or autotrophs) such as plants and algae that convert energy and inorganic material into organic compounds, which can then be used by the rest of the community.[53][260][261] At the next level are the heterotrophs, which are the species that obtain energy by breaking apart organic compounds from other organisms.[259] Heterotrophs that consume plants are primary consumers (or herbivores) whereas heterotrophs that consume herbivores are secondary consumers (or carnivores). And those that eat secondary consumers are tertiary consumers and so on. Omnivorous heterotrophs are able to consume at multiple levels. Finally, there are decomposers that feed on the waste products or dead bodies of organisms.[259]
On average, the total amount of energy incorporated into the biomass of a trophic level per unit of time is about one-tenth of the energy of the trophic level that it consumes. Waste and dead material used by decomposers as well as heat lost from metabolism make up the other ninety percent of energy that is not consumed by the next trophic level.[262]
Biosphere

In the global ecosystem (or biosphere), matter exist as different interacting compartments, which can be biotic or abiotic as well as accessible or inaccessible, depending on their forms and locations.[264] For example, matter from terrestrial autotrophs are both biotic and accessible to other organisms whereas the matter in rocks and minerals are abiotic and inaccessible. A biogeochemical cycle is a pathway by which specific elements of matter are turned over or moved through the biotic (biosphere) and the abiotic (lithosphere, atmosphere, and hydrosphere) compartments of Earth. There are biogeochemical cycles for nitrogen, carbon, and water. In some cycles there are reservoirs where a substance remains or is sequestered for a long period of time.
Climate change includes both global warming driven by human-induced emissions of greenhouse gases and the resulting large-scale shifts in weather patterns. Though there have been previous periods of climatic change, since the mid-20th century humans have had an unprecedented impact on Earth's climate system and caused change on a global scale.[265] The largest driver of warming is the emission of greenhouse gases, of which more than 90% are carbon dioxide and methane.[266] Fossil fuel burning (coal, oil, and natural gas) for energy consumption is the main source of these emissions, with additional contributions from agriculture, deforestation, and manufacturing.[267] Temperature rise is accelerated or tempered by climate feedbacks, such as loss of sunlight-reflecting snow and ice cover, increased water vapor (a greenhouse gas itself), and changes to land and ocean carbon sinks.
Conservation
Conservation biology is the study of the conservation of Earth's biodiversity with the aim of protecting species, their habitats, and ecosystems from excessive rates of extinction and the erosion of biotic interactions.[268][269][270] It is concerned with factors that influence the maintenance, loss, and restoration of biodiversity and the science of sustaining evolutionary processes that engender genetic, population, species, and ecosystem diversity.[271][272][273][274] The concern stems from estimates suggesting that up to 50% of all species on the planet will disappear within the next 50 years,[275] which has contributed to poverty, starvation, and will reset the course of evolution on this planet.[276][277] Biodiversity affects the functioning of ecosystems, which provide a variety of services upon which people depend.
Conservation biologists research and educate on the trends of biodiversity loss, species extinctions, and the negative effect these are having on our capabilities to sustain the well-being of human society. Organizations and citizens are responding to the current biodiversity crisis through conservation action plans that direct research, monitoring, and education programs that engage concerns at local through global scales.[278][271][272][273]
See also
- Biology in fiction
- Glossary of biology
- List of biological websites
- List of biologists
- List of biology journals
- List of biology topics
- List of life sciences
- List of omics topics in biology
- National Association of Biology Teachers
- Outline of biology
- Periodic table of life sciences in Tinbergen's four questions
- Reproduction
- Science tourism
- Terminology of biology
References
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Evolution, the themes of biology, and scientific inquiry". Campbell Biology (11th ed.). New York: Pearson. pp. 2–26. ISBN 978-0134093413.
- Hillis, David M.; Heller, H. Craig; Hacker, Sally D.; Laskowski, Marta J.; Sadava, David E. (2020). "Studying life". Life: The Science of Biology (12th ed.). W. H. Freeman. ISBN 978-1319017644.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Biology and the three of life". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 1–18. ISBN 978-0321976499.
- Modell, Harold; Cliff, William; Michael, Joel; McFarland, Jenny; Wenderoth, Mary Pat; Wright, Ann (December 2015). "A physiologist's view of homeostasis". Advances in Physiology Education. 39 (4): 259–266. doi:10.1152/advan.00107.2015. ISSN 1043-4046. PMC 4669363. PMID 26628646.
- Davies, PC; Rieper, E; Tuszynski, JA (January 2013). "Self-organization and entropy reduction in a living cell". Bio Systems. 111 (1): 1–10. doi:10.1016/j.biosystems.2012.10.005. PMC 3712629. PMID 23159919.
- Based on definition from: "Aquarena Wetlands Project glossary of terms". Texas State University at San Marcos. Archived from the original on 2004-06-08.
- Craig, Nancy (2014). Molecular Biology, Principles of Genome Function. ISBN 978-0-19-965857-2.
- Mosconi, Francesco; Julou, Thomas; Desprat, Nicolas; Sinha, Deepak Kumar; Allemand, Jean-François; Vincent Croquette; Bensimon, David (2008). "Some nonlinear challenges in biology". Nonlinearity. 21 (8): T131. Bibcode:2008Nonli..21..131M. doi:10.1088/0951-7715/21/8/T03. ISSN 0951-7715. S2CID 119808230.
- Howell, Elizabeth (8 December 2014). "How Did Life Become Complex, And Could It Happen Beyond Earth?". Astrobiology Magazine. Archived from the original on 17 August 2018. Retrieved 14 February 2018.
- Pearce, Ben K.D.; Tupper, Andrew S.; Pudritz, Ralph E.; et al. (March 1, 2018). "Constraining the Time Interval for the Origin of Life on Earth". Astrobiology. 18 (3): 343–364. arXiv:1808.09460. Bibcode:2018AsBio..18..343P. doi:10.1089/ast.2017.1674. ISSN 1531-1074. PMID 29570409. S2CID 4419671.
- Lindberg, David C. (2007). "Science before the Greeks". The beginnings of Western science: the European Scientific tradition in philosophical, religious, and institutional context (Second ed.). Chicago, Illinois: University of Chicago Press. pp. 1–20. ISBN 978-0-226-48205-7.
- Grant, Edward (2007). "Ancient Egypt to Plato". A History of Natural Philosophy: From the Ancient World to the Nineteenth Century (First ed.). New York, New York: Cambridge University Press. pp. 1–26. ISBN 978-052-1-68957-1.
- Magner, Lois N. (2002). A History of the Life Sciences, Revised and Expanded. CRC Press. ISBN 978-0-203-91100-6. Archived from the original on 2015-03-24.
- Serafini, Anthony (2013). The Epic History of Biology. ISBN 978-1-4899-6327-7. Archived from the original on 15 April 2021. Retrieved 14 July 2015.
- One or more of the preceding sentences incorporates text from a publication now in the public domain: Chisholm, Hugh, ed. (1911). "Theophrastus". Encyclopædia Britannica (11th ed.). Cambridge University Press.
- Fahd, Toufic (1996). "Botany and agriculture". In Morelon, Régis; Rashed, Roshdi (eds.). Encyclopedia of the History of Arabic Science. Vol. 3. Routledge. p. 815. ISBN 978-0-415-12410-2.
- Magner, Lois N. (2002). A History of the Life Sciences, Revised and Expanded. CRC Press. pp. 133–44. ISBN 978-0-203-91100-6. Archived from the original on 2015-03-24.
- Sapp, Jan (2003). "7". Genesis: The Evolution of Biology. New York: Oxford University Press. ISBN 978-0-19-515618-8.
- Coleman, William (1977). Biology in the Nineteenth Century: Problems of Form, Function, and Transformation. New York: Cambridge University Press. ISBN 978-0-521-29293-1.
- Mayr, Ernst. The Growth of Biological Thought, chapter 4
- Mayr, Ernst. The Growth of Biological Thought, chapter 7
- Darwin 1909, p. 53
- Gould, Stephen Jay. The Structure of Evolutionary Theory. The Belknap Press of Harvard University Press: Cambridge, 2002. ISBN 0-674-00613-5. p. 187.
- Lamarck (1914)
- Mayr, Ernst. The Growth of Biological Thought, chapter 10: "Darwin's evidence for evolution and common descent"; and chapter 11: "The causation of evolution: natural selection"
- Larson, Edward J. (2006). "Ch. 3". Evolution: The Remarkable History of a Scientific Theory. Random House Publishing Group. ISBN 978-1-58836-538-5. Archived from the original on 2015-03-24.
- Henig (2000). Op. cit. pp. 134–138.
- Miko, Ilona (2008). "Gregor Mendel's principles of inheritance form the cornerstone of modern genetics. So just what are they?". Nature Education. 1 (1): 134. Archived from the original on 2019-07-19. Retrieved 2021-05-13.
- Futuyma, Douglas J.; Kirkpatrick, Mark (2017). "Evolutionary Biology". Evolution (4th ed.). Sunderland, Mass.: Sinauer Associates. pp. 3–26.
- Noble, Ivan (2003-04-14). "Human genome finally complete". BBC News. Archived from the original on 2006-06-14. Retrieved 2006-07-22.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "The chemical context of life". Campbell Biology (11th ed.). New York: Pearson. pp. 28–43. ISBN 978-0134093413.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Water and carbon: The chemical basis of life". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 55–77. ISBN 978-0321976499.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Carbon and the molecular diversity of life". Campbell Biology (11th ed.). New York: Pearson. pp. 56–65. ISBN 978-0134093413.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Carbon and molecular diversity of life". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 56–65. ISBN 978-1464175121.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Protein structure and function". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 78–92. ISBN 978-0321976499.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "The structure and function of large biological molecules". Campbell Biology (11th ed.). New York: Pearson. pp. 66–92. ISBN 978-0134093413.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "An introduction to carbohydrate". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 107–118. ISBN 978-0321976499.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Lipids, membranes, and the first cells". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 119–141. ISBN 978-0321976499.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Nucleic acids and the RNA world". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 93–106. ISBN 978-0321976499.
- Mazzarello, P (May 1999). "A unifying concept: the history of cell theory". Nature Cell Biology. 1 (1): E13–15. doi:10.1038/8964. PMID 10559875. S2CID 7338204.
- Campbell NA, Williamson B, Heyden RJ (2006). Biology: Exploring Life. Boston: Pearson Prentice Hall. ISBN 9780132508827. Archived from the original on 2014-11-02. Retrieved 2021-05-13.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Membrane structure and function". Campbell Biology (11th ed.). New York: Pearson. pp. 126–142. ISBN 978-0134093413.
- Alberts B, Johnson A, Lewis J, et al. (2002). Molecular Biology of the Cell (4th ed.). New York: Garland Science. ISBN 978-0-8153-3218-3. Archived from the original on 2017-12-20.
- Tom Herrmann; Sandeep Sharma (March 2, 2019). "Physiology, Membrane". StatPearls. PMID 30855799. Archived from the original on February 17, 2022. Retrieved May 14, 2021.
- Cell Movements and the Shaping of the Vertebrate Body Archived 2020-01-22 at the Wayback Machine in Chapter 21 of Molecular Biology of the Cell Archived 2017-09-27 at the Wayback Machine fourth edition, edited by Bruce Alberts (2002) published by Garland Science.
The Alberts text discusses how the "cellular building blocks" move to shape developing embryos. It is also common to describe small molecules such as amino acids as "molecular building blocks Archived 2020-01-22 at the Wayback Machine". - Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Cells: The working units of life". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 60–81. ISBN 978-1464175121.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Energy and enzymes: An introduction to metabolism". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 171–188. ISBN 978-0321976499.
- Bailey, Regina. "Cellular Respiration". Archived from the original on 2012-05-05.
- Lodish, Harvey; Berk, Arnold.; Kaiser, Chris A.; Krieger, Monty; Scott, Matthew P.; Bretscher, Anthony; Ploegh, Hidde; Matsudaira, Paul (2008). "Cellular energetics". Molecular Cell Biology (6th ed.). New York: W.H. Freeman and Company. pp. 479–532. ISBN 978-0716776017.
- "photosynthesis". Online Etymology Dictionary. Archived from the original on 2013-03-07. Retrieved 2013-05-23.
- φῶς. Liddell, Henry George; Scott, Robert; A Greek–English Lexicon at the Perseus Project
- σύνθεσις. Liddell, Henry George; Scott, Robert; A Greek–English Lexicon at the Perseus Project
- Bryant DA, Frigaard NU (Nov 2006). "Prokaryotic photosynthesis and phototrophy illuminated". Trends in Microbiology. 14 (11): 488–496. doi:10.1016/j.tim.2006.09.001. PMID 16997562.
- Reece J, Urry L, Cain M, Wasserman S, Minorsky P, Jackson R (2011). Biology (International ed.). Upper Saddle River, N.J.: Pearson Education. pp. 235, 244. ISBN 978-0-321-73975-9.
This initial incorporation of carbon into organic compounds is known as carbon fixation.
- Neitzel, James; Rasband, Matthew. "Cell communication". Nature Education. Archived from the original on 29 September 2010. Retrieved 29 May 2021.
- "Cell signaling". Nature Education. Archived from the original on 31 October 2010. Retrieved 29 May 2021.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Cell membranes and signaling". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 82–104. ISBN 978-1464175121.
- Martin EA, Hine R (2020). A dictionary of biology (6th ed.). Oxford: Oxford University Press. ISBN 9780199204625. OCLC 176818780.
- Griffiths AJ (2012). Introduction to genetic analysis (10th ed.). New York: W.H. Freeman and Co. ISBN 9781429229432. OCLC 698085201.
- "10.2 The Cell Cycle - Biology 2e | OpenStax". openstax.org. Archived from the original on 2020-11-29. Retrieved 2020-11-24.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Meiosis". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 271–289. ISBN 978-0321976499.
- Casiraghi A, Suigo L, Valoti E, Straniero V (February 2020). "Targeting Bacterial Cell Division: A Binding Site-Centered Approach to the Most Promising Inhibitors of the Essential Protein FtsZ". Antibiotics. 9 (2): 69. doi:10.3390/antibiotics9020069. PMC 7167804. PMID 32046082.
- Griffiths, Anthony J.; Wessler, Susan R.; Carroll, Sean B.; Doebley, John (2015). "The genetics revolution". An Introduction to Genetic Analysis (11th ed.). Sunderland, Mass.: W.H. Freeman & Company. pp. 1–30. ISBN 978-1464109485.
- Griffiths, Anthony J.F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M., eds. (2000). "Genetics and the Organism: Introduction". An Introduction to Genetic Analysis (7th ed.). New York: W. H. Freeman. ISBN 978-0-7167-3520-5.
- Hartl, D; Jones, E (2005). Genetics: Analysis of Genes and Genomes (6th ed.). Jones & Bartlett. ISBN 978-0-7637-1511-3.
- Rutgers: Mendelian Principles Archived 2021-05-14 at the Wayback Machine
- Miko, Ilona (2008), "Test crosses", Nature Education, 1 (1): 136, archived from the original on 2021-05-21, retrieved 2021-05-28
- Miko, Ilona (2008), "Thomas Hunt Morgan and sex linkage", Nature Education, 1 (1): 143, archived from the original on 2021-05-20, retrieved 2021-05-28
- "Pedigree". National Human Genome Research Institute. Archived from the original on 16 June 2021. Retrieved 28 May 2021.
A pedigree is a genetic representation of a family tree that diagrams the inheritance of a trait or disease though several generations. The pedigree shows the relationships between family members and indicates which individuals express or silently carry the trait in question.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Mendel and the gene idea". Campbell Biology (11th ed.). New York: Pearson. pp. 269–293. ISBN 978-0134093413.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "DNA and its role in heredity". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 172–193. ISBN 978-1464175121.
- Russell P (2001). iGenetics. New York: Benjamin Cummings. ISBN 0-8053-4553-1.
- Thanbichler, M; Wang, SC; Shapiro, L (October 2005). "The bacterial nucleoid: a highly organized and dynamic structure". Journal of Cellular Biochemistry. 96 (3): 506–21. doi:10.1002/jcb.20519. PMID 15988757. S2CID 25355087.
- "Genotype definition – Medical Dictionary definitions". Medterms.com. 2012-03-19. Archived from the original on 2013-09-21. Retrieved 2013-10-02.
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2014). Molecular Biology of the Cell (6th ed.). Garland. p. Chapter 4: DNA, Chromosomes and Genomes. ISBN 978-0-8153-4432-2. Archived from the original on 14 July 2014.
- Purcell A. "DNA". Basic Biology. Archived from the original on 5 January 2017.
- McCarthy D, Minner C, Bernstein H, Bernstein C (October 1976). "DNA elongation rates and growing point distributions of wild-type phage T4 and a DNA-delay amber mutant". Journal of Molecular Biology. 106 (4): 963–81. doi:10.1016/0022-2836(76)90346-6. PMID 789903
- Crick FH (1958). "On protein synthesis". Symposia of the Society for Experimental Biology. 12: 138–63. PMID 13580867.
- Crick F (August 1970). "Central dogma of molecular biology". Nature. 227 (5258): 561–3. Bibcode:1970Natur.227..561C. doi:10.1038/227561a0. PMID 4913914. S2CID 4164029.
- "Central dogma reversed". Nature. 226 (5252): 1198–9. June 1970. Bibcode:1970Natur.226.1198.. doi:10.1038/2261198a0. PMID 5422595. S2CID 4184060.
- Lin, Yihan; Elowitz, Michael B. (2016). "Central Dogma Goes Digital". Molecular Cell. 61 (6): 791–792. doi:10.1016/j.molcel.2016.03.005. PMID 26990983. Archived from the original on 2021-10-03. Retrieved 2021-10-03.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "From DNA to protein: Gene expression". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 194–214. ISBN 978-1464175121.
- "Uracil". Genome.gov. Archived from the original on 2019-10-19. Retrieved 2019-11-21.
- Temin HM, Mizutani S (June 1970). "RNA-dependent DNA polymerase in virions of Rous sarcoma virus". Nature. 226 (5252): 1211–3. doi:10.1038/2261211a0. PMID 4316301. S2CID 4187764.
- Baltimore D (June 1970). "RNA-dependent DNA polymerase in virions of RNA tumour viruses". Nature. 226 (5252): 1209–11. doi:10.1038/2261209a0. PMID 4316300. S2CID 4222378.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Regulation of gene expression". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 215–233. ISBN 978-1464175121.
- Keene, Jack D.; Tenenbaum, Scott A. (2002). "Eukaryotic mRNPs may represent posttranscriptional operons". Molecular Cell. 9 (6): 1161–1167. doi:10.1016/s1097-2765(02)00559-2. PMID 12086614.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Control of gene expression in eukaryotes". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 379–397. ISBN 978-0321976499.
- "WHO definitions of genetics and genomics". World Health Organization. Archived from the original on June 30, 2004.
- Concepts of genetics (10th ed.). San Francisco: Pearson Education. 2012. ISBN 978-0-321-72412-0.
- Culver KW, Labow MA (8 November 2002). "Genomics". In Robinson R (ed.). Genetics. Macmillan Science Library. Macmillan Reference USA. ISBN 978-0-02-865606-9.
- Kadakkuzha BM, Puthanveettil SV (July 2013). "Genomics and proteomics in solving brain complexity". Molecular BioSystems. 9 (7): 1807–21. doi:10.1039/C3MB25391K. PMC 6425491. PMID 23615871.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Genomes". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 234–252. ISBN 978-1464175121.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Biotechnology". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 253–272. ISBN 978-1464175121.
- Griffiths, Anthony J.; Wessler, Susan R.; Carroll, Sean B.; Doebley, John (2015). "Gene isolation and manipulation". An Introduction to Genetic Analysis (11th ed.). Sunderland, Mass.: W.H. Freeman & Company. pp. 351–395. ISBN 978-1464109485.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Biology and the three of life". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 398–417. ISBN 978-0321976499.
- Knabe J.F.; et al. (2008). Evolution and Morphogenesis of Differentiated Multicellular Organisms: Autonomously Generated Diffusion Gradients for Positional Information. Artificial Life XI: Proceedings of the Eleventh International Conference on the Simulation and Synthesis of Living Systems. Archived from the original on 2018-09-13. Retrieved 2021-10-04.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Genes, development, and evolution". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 273–298. ISBN 978-1464175121.
- Slack, J.M.W. (2013) Essential Developmental Biology. Wiley-Blackwell, Oxford.
- Slack, J.M.W. (2007). "Metaplasia and transdifferentiation: from pure biology to the clinic". Nature Reviews Molecular Cell Biology. 8 (5): 369–378. doi:10.1038/nrm2146. PMID 17377526. S2CID 3353748.
- Atala A, Lanza R (2012-12-31). Handbook of Stem Cells. Academic Press. p. 452. ISBN 978-0-12-385943-3. Archived from the original on 2021-04-12. Retrieved 2021-05-28.
- Yanes, Oscar; Clark, Julie; Wong, Diana M.; Patti, Gary J.; Sánchez-Ruiz, Antonio; Benton, H. Paul; Trauger, Sunia A.; Desponts, Caroline; Ding, Sheng; Siuzdak, Gary (June 2010). "Metabolic oxidation regulates embryonic stem cell differentiation". Nature Chemical Biology. 6 (6): 411–417. doi:10.1038/nchembio.364. ISSN 1552-4469. PMC 2873061. PMID 20436487.
- Carroll, Sean B. "The Origins of Form". Natural History. Archived from the original on 9 October 2018. Retrieved 9 October 2016.
Biologists could say, with confidence, that forms change, and that natural selection is an important force for change. Yet they could say nothing about how that change is accomplished. How bodies or body parts change, or how new structures arise, remained complete mysteries.
- Abzhanov, A.; Protas, M.; Grant, B.R.; Grant, P.R.; Tabin, C.J. (2004). "Bmp4 and Morphological Variation of Beaks in Darwin's Finches". Science. 305 (5689): 1462–1465. Bibcode:2004Sci...305.1462A. doi:10.1126/science.1098095. PMID 15353802. S2CID 17226774.
- Cohn, M.J.; Tickle, C. (1999). "Developmental basis of limblessness and axial patterning in snakes". Nature. 399 (6735): 474–479. Bibcode:1999Natur.399..474C. doi:10.1038/20944. PMID 10365960. S2CID 4309833.
- Beverdam, A.; Merlo, G.R.; Paleari, L.; Mantero, S.; Genova, F.; Barbieri, O.; Janvier, P.; Levi, G. (August 2002). "Jaw Transformation With Gain of Symmetry After DLX5/DLX6 Inactivation: Mirror of the Past?" (PDF). Genesis. 34 (4): 221–227. doi:10.1002/gene.10156. hdl:2318/87307. PMID 12434331. S2CID 19592597. Archived (PDF) from the original on 2022-07-30. Retrieved 2021-05-28.
- Depew, M.J.; Lufkin, T.; Rubenstein, J.L. (October 2002). "Specification of jaw subdivisions by DLX genes". Science. 298 (5592): 381–385. doi:10.1126/science.1075703. PMID 12193642. S2CID 10274300.
- Panganiban, Grace; Rubenstein, John L. R. (2002). "Developmental functions of the Distal-less/Dlx homeobox genes". Development. 129 (19): 4371–4386. doi:10.1242/dev.129.19.4371. PMID 12223397. Archived from the original on 2020-08-10. Retrieved 2021-05-28.
- Beldade, P.; Brakefield, P.M.; Long, A.D. (2002). "Contribution of Distal-less to quantitative variation in butterfly eyespots". Nature. 415 (6869): 315–318. doi:10.1038/415315a. PMID 11797007. S2CID 4430563.
- Hall & Hallgrímsson 2007, pp. 4–6
- "Evolution Resources". Washington, D.C.: National Academies of Sciences, Engineering, and Medicine. 2016. Archived from the original on 2016-06-03.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Processes of evolution". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 299–324. ISBN 978-1464175121.
- Packard, Alpheus Spring (1901). Lamarck, the founder of Evolution: his life and work with translations of his writings on organic evolution. New York: Longmans, Green. ISBN 978-0-405-12562-1.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Descent with modifications: A Darwinian view of life". Campbell Biology (11th ed.). New York: Pearson. pp. 466–483. ISBN 978-0134093413.
- "The Complete Works of Darwin Online – Biography". darwin-online.org.uk. Archived from the original on 2007-01-07. Retrieved 2006-12-15.
- Dobzhansky, T. (1973). "Nothing in biology makes sense except in the light of evolution". The American Biology Teacher. 35 (3): 125–29. CiteSeerX 10.1.1.525.3586. doi:10.2307/4444260. JSTOR 4444260. S2CID 207358177. - see also Nothing in Biology Makes Sense Except in the Light of Evolution
- Carroll, Joseph, ed. (2003). On the origin of species by means of natural selection. Peterborough, Ontario: Broadview. p. 15. ISBN 978-1-55111-337-1.
As Darwinian scholar Joseph Carroll of the University of Missouri–St. Louis puts it in his introduction to a modern reprint of Darwin's work: "The Origin of Species has special claims on our attention. It is one of the two or three most significant works of all time—one of those works that fundamentally and permanently alter our vision of the world ... It is argued with a singularly rigorous consistency but it is also eloquent, imaginatively evocative, and rhetorically compelling."
- Shermer p. 149.
- Lewontin, Richard C. (November 1970). "The Units of Selection" (PDF). Annual Review of Ecology and Systematics. 1: 1–18. doi:10.1146/annurev.es.01.110170.000245. ISSN 1545-2069. JSTOR 2096764. Archived (PDF) from the original on 2015-02-06.
- Darwin, Charles (1859). On the Origin of Species, John Murray.
- Futuyma, Douglas J.; Kirkpatrick, Mark (2017). "Evolutionary biology". Evolution (4th ed.). Sunderland, Mass.: Sinauer Associates. pp. 3–26.
- Charlesworth, Brian; Charlesworth, Deborah (2009). "Darwin and genetics". Genetics. 183 (3): 757–766. doi:10.1534/genetics.109.109991. PMC 2778973. PMID 19933231.
- Futuyma, Douglas J.; Kirkpatrick, Mark (2017). "Mutation and variation". Evolution (4th ed.). Sunderland, Mass.: Sinauer Associates. pp. 79–101.
- Simpson, George Gaylord (1967). The Meaning of Evolution (Second ed.). Yale University Press. ISBN 978-0-300-00952-1.
- Masel, Joanna (October 25, 2011). "Genetic drift". Current Biology. 21 (20): R837–R838. doi:10.1016/j.cub.2011.08.007. ISSN 0960-9822. PMID 22032182. S2CID 17619958.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Speciation". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 343–356. ISBN 978-1464175121.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Reconstructing and using phylogenies". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 325–342. ISBN 978-1464175121.
- Kitching, Ian J.; Forey, Peter L.; Williams, David M. (2001). "Cladistics". In Levin, Simon A. (ed.). Encyclopedia of Biodiversity (2nd ed.). Elsevier. pp. 33–45. doi:10.1016/B978-0-12-384719-5.00022-8. ISBN 9780123847201. Archived from the original on 29 August 2021. Retrieved 29 August 2021.)
- Futuyma, Douglas J.; Kirkpatrick, Mark (2017). "Phylogeny: The unity and diversity of life". Evolution (4th ed.). Sunderland, Mass.: Sinauer Associates. pp. 401–429.
- Woese, CR; Kandler, O; Wheelis, ML (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–79. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- McNeill, J; Barrie, FR; Buck, WR; Demoulin, V; Greuter, W; Hawksworth, DL; et al. (2012). International Code of Nomenclature for algae, fungi and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. A.R.G. Gantner Verlag KG. ISBN 978-3-87429-425-6. Archived from the original on 2013-11-04. Recommendation 60F
- Silyn-Roberts, Heather (2000). Writing for Science and Engineering: Papers, Presentation. Oxford: Butterworth-Heinemann. p. 198. ISBN 978-0-7506-4636-9. Archived from the original on 2020-10-02. Retrieved 2020-08-24.
- Montévil, M; Mossio, M; Pocheville, A; Longo, G (October 2016). "Theoretical principles for biology: Variation". Progress in Biophysics and Molecular Biology. From the Century of the Genome to the Century of the Organism: New Theoretical Approaches. 122 (1): 36–50. doi:10.1016/j.pbiomolbio.2016.08.005. PMID 27530930. S2CID 3671068. Archived from the original on 2018-03-20.
- De Duve, Christian (2002). Life Evolving: Molecules, Mind, and Meaning. New York: Oxford University Press. p. 44. ISBN 978-0-19-515605-8.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The history of life on Earth". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 357–376. ISBN 978-1464175121.
- "Stratigraphic Chart 2022" (PDF). International Stratigraphic Commission. February 2022. Archived (PDF) from the original on 2 April 2022. Retrieved 25 April 2022.
- Futuyma 2005
- Futuyma, DJ (2005). Evolution. Sinauer Associates. ISBN 978-0-87893-187-3. OCLC 57311264.
- Rosing, Minik T. (January 29, 1999). "13C-Depleted Carbon Microparticles in >3700-Ma Sea-Floor Sedimentary Rocks from West Greenland". Science. 283 (5402): 674–676. Bibcode:1999Sci...283..674R. doi:10.1126/science.283.5402.674. ISSN 0036-8075. PMID 9924024.
- Ohtomo, Yoko; Kakegawa, Takeshi; Ishida, Akizumi; et al. (January 2014). "Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks". Nature Geoscience. 7 (1): 25–28. Bibcode:2014NatGe...7...25O. doi:10.1038/ngeo2025. ISSN 1752-0894.
- Nisbet, Euan G.; Fowler, C.M.R. (December 7, 1999). "Archaean metabolic evolution of microbial mats". Proceedings of the Royal Society B. 266 (1436): 2375–2382. doi:10.1098/rspb.1999.0934. ISSN 0962-8452. PMC 1690475.
- Knoll, Andrew H.; Javaux, Emmanuelle J.; Hewitt, David; et al. (June 29, 2006). "Eukaryotic organisms in Proterozoic oceans". Philosophical Transactions of the Royal Society B. 361 (1470): 1023–1038. doi:10.1098/rstb.2006.1843. ISSN 0962-8436. PMC 1578724. PMID 16754612.
- Fedonkin, Mikhail A. (March 31, 2003). "The origin of the Metazoa in the light of the Proterozoic fossil record" (PDF). Paleontological Research. 7 (1): 9–41. doi:10.2517/prpsj.7.9. ISSN 1342-8144. S2CID 55178329. Archived from the original (PDF) on 2009-02-26. Retrieved 2008-09-02.
- Bonner, John Tyler (January 7, 1998). "The origins of multicellularity". Integrative Biology. 1 (1): 27–36. doi:10.1002/(SICI)1520-6602(1998)1:1<27::AID-INBI4>3.0.CO;2-6. ISSN 1757-9694.
- Strother, Paul K.; Battison, Leila; Brasier, Martin D.; et al. (May 26, 2011). "Earth's earliest non-marine eukaryotes". Nature. 473 (7348): 505–509. Bibcode:2011Natur.473..505S. doi:10.1038/nature09943. ISSN 0028-0836. PMID 21490597. S2CID 4418860.
- Beraldi-Campesi, Hugo (February 23, 2013). "Early life on land and the first terrestrial ecosystems". Ecological Processes. 2 (1): 1–17. doi:10.1186/2192-1709-2-1. ISSN 2192-1709.
- Algeo, Thomas J.; Scheckler, Stephen E. (January 29, 1998). "Terrestrial-marine teleconnections in the Devonian: links between the evolution of land plants, weathering processes, and marine anoxic events". Philosophical Transactions of the Royal Society B. 353 (1365): 113–130. doi:10.1098/rstb.1998.0195. ISSN 0962-8436. PMC 1692181.
- Jun-Yuan, Chen; Oliveri, Paola; Chia-Wei, Li; et al. (April 25, 2000). "Precambrian animal diversity: Putative phosphatized embryos from the Doushantuo Formation of China". Proc. Natl. Acad. Sci. U.S.A. 97 (9): 4457–4462. Bibcode:2000PNAS...97.4457C. doi:10.1073/pnas.97.9.4457. ISSN 0027-8424. PMC 18256. PMID 10781044.
- D-G., Shu; H-L., Luo; Conway Morris, Simon; et al. (November 4, 1999). "Lower Cambrian vertebrates from south China" (PDF). Nature. 402 (6757): 42–46. Bibcode:1999Natur.402...42S. doi:10.1038/46965. ISSN 0028-0836. S2CID 4402854. Archived from the original (PDF) on 2009-02-26. Retrieved 2015-01-22.
- Hoyt, Donald F. (February 17, 1997). "Synapsid Reptiles". ZOO 138 Vertebrate Zoology (Lecture). Pomona, Calif.: California State Polytechnic University, Pomona. Archived from the original on 2009-05-20. Retrieved 2015-01-22.
- Barry, Patrick L. (January 28, 2002). Phillips, Tony (ed.). "The Great Dying". Science@NASA. Marshall Space Flight Center. Archived from the original on 2010-04-10. Retrieved 2015-01-22.
- Tanner, Lawrence H.; Lucas, Spencer G.; Chapman, Mary G. (March 2004). "Assessing the record and causes of Late Triassic extinctions" (PDF). Earth-Science Reviews. 65 (1–2): 103–139. Bibcode:2004ESRv...65..103T. doi:10.1016/S0012-8252(03)00082-5. Archived from the original (PDF) on 2007-10-25. Retrieved 2007-10-22.
- Benton 1997
- Fastovsky, David E.; Sheehan, Peter M. (March 2005). "The Extinction of the Dinosaurs in North America" (PDF). GSA Today. 15 (3): 4–10. doi:10.1130/1052-5173(2005)015<4:TEOTDI>2.0.CO;2. ISSN 1052-5173. Archived (PDF) from the original on 2019-03-22. Retrieved 2015-01-23.
- Roach, John (June 20, 2007). "Dinosaur Extinction Spurred Rise of Modern Mammals". National Geographic News. Washington, D.C.: National Geographic Society. Archived from the original on 2008-05-11. Retrieved 2020-02-21.
- Wible, John R.; Rougier, Guillermo W.; Novacek, Michael J.; et al. (June 21, 2007). "Cretaceous eutherians and Laurasian origin for placental mammals near the K/T boundary". Nature. 447 (7147): 1003–1006. Bibcode:2007Natur.447.1003W. doi:10.1038/nature05854. ISSN 0028-0836. PMID 17581585. S2CID 4334424.
- Van Valkenburgh, Blaire (May 1, 1999). "Major Patterns in the History of Carnivorous Mammals". Annual Review of Earth and Planetary Sciences. 27: 463–493. Bibcode:1999AREPS..27..463V. doi:10.1146/annurev.earth.27.1.463. ISSN 1545-4495. Archived from the original on February 29, 2020. Retrieved May 15, 2021.
- Fredrickson JK, Zachara JM, Balkwill DL, Kennedy D, Li SM, Kostandarithes HM, Daly MJ, Romine MF, Brockman FJ (July 2004). "Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington state". Applied and Environmental Microbiology. 70 (7): 4230–41. Bibcode:2004ApEnM..70.4230F. doi:10.1128/AEM.70.7.4230-4241.2004. PMC 444790. PMID 15240306.
- Dudek NK, Sun CL, Burstein D (2017). "Novel Microbial Diversity and Functional Potential in the Marine Mammal Oral Microbiome" (PDF). Current Biology. 27 (24): 3752–3762. doi:10.1016/j.cub.2017.10.040. PMID 29153320. S2CID 43864355. Archived (PDF) from the original on 2021-03-08. Retrieved 2021-05-14.
- Pace NR (May 2006). "Time for a change". Nature. 441 (7091): 289. Bibcode:2006Natur.441..289P. doi:10.1038/441289a. PMID 16710401. S2CID 4431143.
- Stoeckenius W (October 1981). "Walsby's square bacterium: fine structure of an orthogonal procaryote". Journal of Bacteriology. 148 (1): 352–60. doi:10.1128/JB.148.1.352-360.1981. PMC 216199. PMID 7287626.
- "Archaea Basic Biology". March 2018. Archived from the original on 2021-04-28. Retrieved 2021-05-14.
- Bang C, Schmitz RA (September 2015). "Archaea associated with human surfaces: not to be underestimated". FEMS Microbiology Reviews. 39 (5): 631–48. doi:10.1093/femsre/fuv010. PMID 25907112.
- Moissl-Eichinger C, Pausan M, Taffner J, Berg G, Bang C, Schmitz RA (January 2018). "Archaea Are Interactive Components of Complex Microbiomes". Trends in Microbiology. 26 (1): 70–85. doi:10.1016/j.tim.2017.07.004. PMID 28826642.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The origin and diversification of eukaryotes". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 402–419. ISBN 978-1464175121.
- O'Malley, Maureen A.; Leger, Michelle M.; Wideman, Jeremy G.; Ruiz-Trillo, Iñaki (2019-02-18). "Concepts of the last eukaryotic common ancestor". Nature Ecology & Evolution. Springer Science and Business Media LLC. 3 (3): 338–344. doi:10.1038/s41559-019-0796-3. hdl:10261/201794. ISSN 2397-334X. PMID 30778187. S2CID 67790751.
- Taylor, F. J. R. 'M. (2003-11-01). "The collapse of the two-kingdom system, the rise of protistology and the founding of the International Society for Evolutionary Protistology (ISEP)". International Journal of Systematic and Evolutionary Microbiology. Microbiology Society. 53 (6): 1707–1714. doi:10.1099/ijs.0.02587-0. ISSN 1466-5026. PMID 14657097.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The evolution of plants". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 420–449. ISBN 978-1464175121.
- "Gymnosperms on The Plant List". Theplantlist.org. Archived from the original on 2013-08-24. Retrieved 2013-07-24.
- Christenhusz, M. J. M. & Byng, J. W. (2016). "The number of known plants species in the world and its annual increase". Phytotaxa. 261 (3): 201–217. doi:10.11646/phytotaxa.261.3.1. Archived from the original on 2016-07-29. Retrieved 2021-05-15.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The evolution and diversity of fungi". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 451–468. ISBN 978-1464175121.
- Wallen, R. M.; Perlin, Michael H. (2018-03-21). "An Overview of the Function and Maintenance of Sexual Reproduction in Dikaryotic Fungi". Frontiers in Microbiology. 9: 503. doi:10.3389/fmicb.2018.00503. PMC 5871698. PMID 29619017.
- Hawksworth DL, Lücking R (July 2017). "Fungal Diversity Revisited: 2.2 to 3.8 Million Species". The Fungal Kingdom. Microbiology Spectrum. Vol. 5. pp. 79–95. doi:10.1128/microbiolspec.FUNK-0052-2016. ISBN 978-1-55581-957-6. PMID 28752818.
- Cheek, Martin; Nic Lughadha, Eimear; Kirk, Paul; Lindon, Heather; Carretero, Julia; Looney, Brian; et al. (2020). "New scientific discoveries: Plants and fungi". Plants, People, Planet. 2 (5): 371–388. doi:10.1002/ppp3.10148.
- "Stop neglecting fungi". Nature Microbiology. 2 (8): 17120. 25 July 2017. doi:10.1038/nmicrobiol.2017.120. PMID 28741610.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Animal origins and diversity". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 469–519. ISBN 978-1464175121.
- Feuda R, Dohrmann M, Pett W, Philippe H, Rota-Stabelli O, Lartillot N, et al. (December 2017). "Improved Modeling of Compositional Heterogeneity Supports Sponges as Sister to All Other Animals". Current Biology. 27 (24): 3864–3870.e4. doi:10.1016/j.cub.2017.11.008. PMID 29199080.
- Pisani D, Pett W, Dohrmann M, Feuda R, Rota-Stabelli O, Philippe H, et al. (December 2015). "Genomic data do not support comb jellies as the sister group to all other animals". Proceedings of the National Academy of Sciences of the United States of America. 112 (50): 15402–7. Bibcode:2015PNAS..11215402P. doi:10.1073/pnas.1518127112. PMC 4687580. PMID 26621703.
- Simion P, Philippe H, Baurain D, Jager M, Richter DJ, Di Franco A, et al. (April 2017). "A Large and Consistent Phylogenomic Dataset Supports Sponges as the Sister Group to All Other Animals" (PDF). Current Biology. 27 (7): 958–967. doi:10.1016/j.cub.2017.02.031. PMID 28318975. Archived (PDF) from the original on 2020-04-25. Retrieved 2021-05-14.
- Giribet G (1 October 2016). "Genomics and the animal tree of life: conflicts and future prospects". Zoologica Scripta. 45: 14–21. doi:10.1111/zsc.12215. ISSN 1463-6409.
- Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, Giribet G (2017-10-11). "Placozoans are eumetazoans related to Cnidaria". bioRxiv 10.1101/200972.
- May, Robert M. (16 September 1988). "How Many Species Are There on Earth?". Science. 241 (4872): 1441–1449. Bibcode:1988Sci...241.1441M. doi:10.1126/science.241.4872.1441. JSTOR 1702670. PMID 17790039. S2CID 34992724. Archived from the original on 15 November 2016. Retrieved 17 June 2014.
- Richards, O. W.; Davies, R.G. (1977). Imms' General Textbook of Entomology: Volume 1: Structure, Physiology and Development Volume 2: Classification and Biology. Berlin: Springer. ISBN 978-0-412-61390-6.
- "Table 1a: Number of species evaluated in relation to the overall number of described species, and numbers of threatened species by major groups of organisms". IUCN Red List. 18 July 2019. Archived from the original on 20 December 2020. Retrieved 14 May 2021.
- Wu KJ (15 April 2020). "There are more viruses than stars in the universe. Why do only some infect us? – More than a quadrillion quadrillion individual viruses exist on Earth, but most are not poised to hop into humans. Can we find the ones that are?". National Geographic Society. Archived from the original on 28 May 2020. Retrieved 18 May 2020.
- Koonin EV, Senkevich TG, Dolja VV (September 2006). "The ancient Virus World and evolution of cells". Biology Direct. 1 (1): 29. doi:10.1186/1745-6150-1-29. PMC 1594570. PMID 16984643.
- Zimmer C (26 February 2021). "The Secret Life of a Coronavirus - An oily, 100-nanometer-wide bubble of genes has killed more than two million people and reshaped the world. Scientists don't quite know what to make of it". The New York Times. Archived from the original on 2021-12-28. Retrieved 28 February 2021.
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Archived from the original on 20 March 2020. Retrieved 25 April 2020.
- Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ (May 2009). "Structural and functional studies of archaeal viruses". The Journal of Biological Chemistry. 284 (19): 12599–603. doi:10.1074/jbc.R800078200. PMC 2675988. PMID 19158076.
- Edwards RA, Rohwer F (June 2005). "Viral metagenomics". Nature Reviews. Microbiology. 3 (6): 504–10. doi:10.1038/nrmicro1163. PMID 15886693. S2CID 8059643.
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H (August 2003). "Phage as agents of lateral gene transfer". Current Opinion in Microbiology. 6 (4): 417–24. doi:10.1016/S1369-5274(03)00086-9. PMID 12941415.
- Rybicki EP (1990). "The classification of organisms at the edge of life, or problems with virus systematics". South African Journal of Science. 86: 182–86.
- Koonin EV, Starokadomskyy P (October 2016). "Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question". Studies in History and Philosophy of Biological and Biomedical Sciences. 59: 125–34. doi:10.1016/j.shpsc.2016.02.016. PMC 5406846. PMID 26965225.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The plant body". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 521–536. ISBN 978-1464175121.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Plant nutrition and transport". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 537–554. ISBN 978-1464175121.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Plant growth and development". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 555–572. ISBN 978-1464175121.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Reproduction of flowering plants". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 573–588. ISBN 978-1464175121.
- "Self-Pollination and Cross-Pollination | Biology for Majors II". courses.lumenlearning.com. Archived from the original on 2020-07-21. Retrieved 2021-05-15.
- Harmer SL, Panda S, Kay SA (2001). "Molecular bases of circadian rhythms". Annual Review of Cell and Developmental Biology. 17: 215–53. doi:10.1146/annurev.cellbio.17.1.215. PMID 11687489.
- Strong, Donald R.; Ray, Thomas S. (1 January 1975). "Host Tree Location Behavior of a Tropical Vine (Monstera gigantea) by Skototropism". Science. 190 (4216): 804–806. Bibcode:1975Sci...190..804S. doi:10.1126/science.190.4216.804. JSTOR 1741614. S2CID 84386403.
- Jaffe MJ, Forbes S (February 1993). "Thigmomorphogenesis: the effect of mechanical perturbation on plants". Plant Growth Regulation. 12 (3): 313–24. doi:10.1007/BF00027213. PMID 11541741. S2CID 29466083.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Fundamentals of animal function". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 605–623. ISBN 978-1464175121.
- Rodolfo, Kelvin (January 2000). "What is homeostasis?". Scientific American. Archived from the original on 2013-12-03.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Water and salt balance". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 751–767. ISBN 978-1464175121.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Nutrition, feeding, and digestion". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 624–642. ISBN 978-1464175121.
- Campbell, Neil A. (1990). Biology (2nd ed.). Redwood City, Calif.: Benjamin/Cummings Pub. Co. pp. 834–835. ISBN 0-8053-1800-3.
- Hsia, CC; Hyde, DM; Weibel, ER (15 March 2016). "Lung Structure and the Intrinsic Challenges of Gas Exchange". Comprehensive Physiology. 6 (2): 827–95. doi:10.1002/cphy.c150028. PMC 5026132. PMID 27065169.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Circulation". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 661–680. ISBN 978-1464175121.
- "cardiovascular system" at Dorland's Medical Dictionary
- "How does the blood circulatory system work?". PubMed Health. 1 August 2016. Archived from the original on 28 August 2021. Retrieved 14 May 2021.
- Pawlina, Wojciech; Ross, Michael H. (2011). Histology : a text and atlas : with correlated cell and molecular biology (6th ed.). Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health. ISBN 9780781772006. OCLC 548651322.
- Standring, Susan (2016). Gray's anatomy : the anatomical basis of clinical practice (Forty-first ed.). Philadelphia. ISBN 9780702052309. OCLC 920806541.
- Hillis, David M.; Sadava, David E.; Price, Mary V. (2014). "Muscle and movement". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 681–698. ISBN 978-1-464-10947-8.
- Gardner, C.R. (1976). "The neuronal control of locomotion in the earthworm". Biological Reviews of the Cambridge Philosophical Society. 51 (1): 25–52. doi:10.1111/j.1469-185X.1976.tb01119.x. PMID 766843. S2CID 9983649.
- Alexander, R. McNeill (2003). "Muscle, the motor". Principles of Animal Locomotion (2nd ed.). Princeton, N.J.: Princeton University Press. pp. 15–37. ISBN 978-0-691-12634-0.
- Josephson, R. K.; Malamud, J. G.; Stokes, D. R. (2000-09-15). "Asynchronous muscle: a primer". Journal of Experimental Biology. 203 (18): 2713–2722. doi:10.1242/jeb.203.18.2713. ISSN 0022-0949. PMID 10952872. Archived from the original on 2020-10-31. Retrieved 2021-05-14.
- Lee WC, Huang H, Feng G, Sanes JR, Brown EN, So PT, Nedivi E (February 2006). "Dynamic remodeling of dendritic arbors in GABAergic interneurons of adult visual cortex". PLOS Biology. 4 (2): e29. doi:10.1371/journal.pbio.0040029. PMC 1318477. PMID 16366735.
- "Nervous System". Columbia Encyclopedia. Columbia University Press.
- Aidley, David J. (1998). "Introduction". The Physiology of Excitable Cells (4th ed.). New York: Cambridge University Press. pp. 1–7. ISBN 978-0521574211.
- Aidley, David J. (1998). "The ionic basis of nervous conduction". The Physiology of Excitable Cells (4th ed.). New York: Cambridge University Press. pp. 54–75. ISBN 978-0521574211.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Neurons, sense organs, and nervous systems". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 699–732. ISBN 978-1464175121.
- Freeman, Scott; Quillin, Kim; Allison, Lizabeth; Black, Michael; Podgorski, Greg; Taylor, Emily; Carmichael, Jeff (2017). "Animal sensory systems". Biological Science (6th ed.). Hoboken, N.J.: Pearson. pp. 922–941. ISBN 978-0321976499.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Control by the endocrine and nervous systems". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 733–750. ISBN 978-1464175121.
- Shuster M (2014-03-14). Biology for a changing world, with physiology (Second ed.). New York, NY. ISBN 9781464151132. OCLC 884499940.
- Marieb E (2014). Anatomy & physiology. Glenview, IL: Pearson Education, Inc. ISBN 978-0-321-86158-0.
- Knobil, Ernst (1998). Encyclopedia of reproduction, Volume 1. Academic Press. p. 315. ISBN 978-0-12-227020-8.
- Schwartz, Jill (2010). Master the GED 2011. Peterson's. p. 371. ISBN 978-0-7689-2885-3.
- Hamilton, Matthew B. (2009). Population genetics. Wiley-Blackwell. p. 55. ISBN 978-1-4051-3277-0.
- Ville, Claude Alvin; Walker, Warren Franklin; Barnes, Robert D. (1984). General zoology. Saunders College Pub. p. 467. ISBN 978-0-03-062451-3.
- Hamilton, William James; Boyd, James Dixon; Mossman, Harland Winfield (1945). Human embryology: (prenatal development of form and function). Williams & Wilkins. p. 330.
- Philips, Joy B. (1975). Development of vertebrate anatomy. Mosby. p. 176. ISBN 978-0-8016-3927-2.
- The Encyclopedia Americana: a library of universal knowledge, Volume 10. Encyclopedia Americana Corp. 1918. p. 281.
- Romoser, William S.; Stoffolano, J. G. (1998). The science of entomology. WCB McGraw-Hill. p. 156. ISBN 978-0-697-22848-2.
- Adiyodi, K.G.; Hughes, Roger N.; Adiyodi, Rita G. (July 2002). Reproductive Biology of Invertebrates, Volume 11, Progress in Asexual Reproduction. Wiley. p. 116. ISBN 978-0-471-48968-9.
- Schatz, Phil. "Concepts of Biology | How Animals Reproduce". OpenStax College. Archived from the original on 6 March 2018. Retrieved 5 March 2018.
- Jungnickel MK, Sutton KA, Florman HM (August 2003). "In the beginning: lessons from fertilization in mice and worms". Cell. 114 (4): 401–4. doi:10.1016/s0092-8674(03)00648-2. PMID 12941269.
- Gilbert, S. F.; Barresi, M. J. F. (2017-05-01). "Developmental Biology, 11Th Edition 2016". American Journal of Medical Genetics Part A. 173 (5): 1430. doi:10.1002/ajmg.a.38166. ISSN 1552-4833.
- Edlund, Helena (July 2002). "Organogenesis: Pancreatic organogenesis — developmental mechanisms and implications for therapy". Nature Reviews Genetics. 3 (7): 524–532. doi:10.1038/nrg841. ISSN 1471-0064. PMID 12094230. S2CID 2436869.
- Rankin, Scott (2018). "Timing is everything: Reiterative Wnt, BMP and RA signaling regulate developmental competence during endoderm organogenesis". Developmental Biology. 434 (1): 121–132. doi:10.1016/j.ydbio.2017.11.018. PMC 5785443. PMID 29217200 – via NCBI.
- Ader, Marius; Tanaka, Elly M (2014). "Modeling human development in 3D culture". Current Opinion in Cell Biology. 31: 23–28. doi:10.1016/j.ceb.2014.06.013. PMID 25033469.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Animal behavior". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 827–844. ISBN 978-1464175121.
- Páez-Rondón, Oscar; Aldana, Elis; Dickens, Joseph; Otálora-Luna, Fernando (May 2018). "Ethological description of a fixed action pattern in a kissing bug (Triatominae): vision, gustation, proboscis extension and drinking of water and guava". Journal of Ethology. 36 (2): 107–116. doi:10.1007/s10164-018-0547-y. ISSN 0289-0771.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Animal behavior". Campbell Biology (11th ed.). New York: Pearson. pp. 1137–1161. ISBN 978-0134093413.
- Begon, M; Townsend, CR; Harper, JL (2006). Ecology: From individuals to ecosystems (4th ed.). Blackwell. ISBN 978-1-4051-1117-1.
- Habitats of the world. New York: Marshall Cavendish. 2004. p. 238. ISBN 978-0-7614-7523-1. Archived from the original on 2021-04-15. Retrieved 2020-08-24.
- Tansley (1934); Molles (1999), p. 482; Chapin et al. (2002), p. 380; Schulze et al. (2005); p. 400; Gurevitch et al. (2006), p. 522; Smith & Smith 2012, p. G-5
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The distribution of Earth's ecological systems". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 845–863. ISBN 978-1464175121.
- Odum, Eugene P (1971). Fundamentals of Ecology (third ed.). New York: Saunders. ISBN 978-0-534-42066-6.
- Chapin III, F. Stuart; Matson, Pamela A.; Mooney, Harold A. (2002). "The ecosystem concept". Principles of Terrestrial Ecosystem Ecology. New York: Springer. p. 10. ISBN 978-0-387-95443-1.
- Planton, Serge, ed. (2013). "Annex III. Glossary: IPCC – Intergovernmental Panel on Climate Change" (PDF). IPCC Fifth Assessment Report. p. 1450. Archived from the original (PDF) on 2016-05-24. Retrieved 25 July 2016.
- Shepherd, Dr. J. Marshall; Shindell, Drew; O'Carroll, Cynthia M. (1 February 2005). "What's the Difference Between Weather and Climate?". NASA. Archived from the original on 22 September 2020. Retrieved 13 November 2015.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Populations". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 864–897. ISBN 978-1464175121.
- Urry, Lisa; Cain, Michael; Wasserman, Steven; Minorsky, Peter; Reece, Jane (2017). "Population ecology". Campbell Biology (11th ed.). New York: Pearson. pp. 1188–1211. ISBN 978-0134093413.
- "Population". Biology Online. Archived from the original on 13 April 2019. Retrieved 5 December 2012.
- "Definition of population (biology)". Oxford Dictionaries. Oxford University Press. Archived from the original on 4 March 2016. Retrieved 5 December 2012.
a community of animals, plants, or humans among whose members interbreeding occurs
- Hartl, Daniel (2007). Principles of Population Genetics. Sinauer Associates. p. 45. ISBN 978-0-87893-308-2.
- Chapman, Eric J.; Byron, Carrie J. (2018-01-01). "The flexible application of carrying capacity in ecology". Global Ecology and Conservation. 13: e00365. doi:10.1016/j.gecco.2017.e00365. ISSN 2351-9894.
- Odum, E. P.; Barrett, G. W. (2005). Fundamentals of Ecology (5th ed.). Brooks/Cole, a part of Cengage Learning. ISBN 978-0-534-42066-6. Archived from the original on 2011-08-20.
- Wootton, JT; Emmerson, M (2005). "Measurement of Interaction Strength in Nature". Annual Review of Ecology, Evolution, and Systematics. 36: 419–44. doi:10.1146/annurev.ecolsys.36.091704.175535. JSTOR 30033811.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Ecological and evolutionary consequences within and among species". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 882–897. ISBN 978-1464175121.
- Smith, AL (1997). Oxford dictionary of biochemistry and molecular biology. Oxford [Oxfordshire]: Oxford University Press. p. 508. ISBN 978-0-19-854768-6.
Photosynthesis – the synthesis by organisms of organic chemical compounds, esp. carbohydrates, from carbon dioxide using energy obtained from light rather than the oxidation of chemical compounds.
- Edwards, Katrina. "Microbiology of a Sediment Pond and the Underlying Young, Cold, Hydrologically Active Ridge Flank". Woods Hole Oceanographic Institution.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Ecological communities". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 898–915. ISBN 978-1464175121.
- Riebeek, Holli (16 June 2011). "The Carbon Cycle". Earth Observatory. NASA. Archived from the original on 5 March 2016. Retrieved 5 April 2018.
- Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "The distribution of Earth's ecological systems". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 916–934. ISBN 978-1464175121.
- IPCC AR5 WG1 Summary for Policymakers 2013, p. 4: Warming of the climate system is unequivocal, and since the 1950s many of the observed changes are unprecedented over decades to millennia. The atmosphere and ocean have warmed, the amounts of snow and ice have diminished, sea level has risen, and the concentrations of greenhouse gases have increased; IPCC SR15 Ch1 2018, p. 54: Abundant empirical evidence of the unprecedented rate and global scale of impact of human influence on the Earth System (Steffen et al., 2016; Waters et al., 2016) has led many scientists to call for an acknowledgment that the Earth has entered a new geological epoch: the Anthropocene.
- EPA 2020: Carbon dioxide (76%), Methane (16%), Nitrous Oxide (6%).
- EPA 2020: Carbon dioxide enters the atmosphere through burning fossil fuels (coal, natural gas, and oil), solid waste, trees and other biological materials, and also as a result of certain chemical reactions (e.g., manufacture of cement). Fossil fuel use is the primary source of CO2. CO2 can also be emitted from direct human-induced impacts on forestry and other land use, such as through deforestation, land clearing for agriculture, and degradation of soils. Methane is emitted during the production and transport of coal, natural gas, and oil. Methane emissions also result from livestock and other agricultural practices and by the decay of organic waste in municipal solid waste landfills.
- Sahney, S.; Benton, M. J (2008). "Recovery from the most profound mass extinction of all time". Proceedings of the Royal Society B: Biological Sciences. 275 (1636): 759–65. doi:10.1098/rspb.2007.1370. PMC 2596898. PMID 18198148.
- Soulé, Michael E.; Wilcox, Bruce A. (1980). Conservation biology: an evolutionary-ecological perspective. Sunderland, Mass.: Sinauer Associates. ISBN 978-0-87893-800-1.
- Soulé, Michael E. (1986). "What is Conservation Biology?" (PDF). BioScience. American Institute of Biological Sciences. 35 (11): 727–34. doi:10.2307/1310054. JSTOR 1310054. Archived from the original (PDF) on 2019-04-12. Retrieved 2021-05-15.
- Hunter, Malcolm L. (1996). Fundamentals of conservation biology. Oxford: Blackwell Science. ISBN 978-0-86542-371-8.
- Meffe, Gary K.; Martha J. Groom (2006). Principles of conservation biology (3rd ed.). Sunderland, Mass.: Sinauer Associates. ISBN 978-0-87893-518-5.
- Van Dyke, Fred (2008). Conservation biology: foundations, concepts, applications (2nd ed.). New York: Springer-Verlag. doi:10.1007/978-1-4020-6891-1. ISBN 9781402068904. OCLC 232001738. Archived from the original on 2020-07-27. Retrieved 2021-05-15.
- Sahney, S.; Benton, M. J.; Ferry, P. A. (2010). "Links between global taxonomic diversity, ecological diversity and the expansion of vertebrates on land". Biology Letters. 6 (4): 544–7. doi:10.1098/rsbl.2009.1024. PMC 2936204. PMID 20106856.
- Koh, Lian Pin; Dunn, Robert R.; Sodhi, Navjot S.; Colwell, Robert K.; Proctor, Heather C.; Smith, Vincent S. (2004). "Species coextinctions and the biodiversity crisis". Science. 305 (5690): 1632–4. Bibcode:2004Sci...305.1632K. doi:10.1126/science.1101101. PMID 15361627. S2CID 30713492.
- Millennium Ecosystem Assessment (2005). Ecosystems and Human Well-being: Biodiversity Synthesis. World Resources Institute, Washington, D.C. Archived 2019-10-14 at the Wayback Machine
- Jackson, J. B. C. (2008). "Ecological extinction and evolution in the brave new ocean". Proceedings of the National Academy of Sciences. 105 (Suppl 1): 11458–65. Bibcode:2008PNAS..10511458J. doi:10.1073/pnas.0802812105. PMC 2556419. PMID 18695220.
- Soule, Michael E. (1986). Conservation Biology: The Science of Scarcity and Diversity. Sinauer Associates. p. 584. ISBN 978-0-87893-795-0.
Further reading
- Benton, Michael J. (1997). Vertebrate Palaeontology (2nd ed.). London: Chapman & Hall. ISBN 978-0-412-73800-5. OCLC 37378512.
- Darwin, Francis, ed. (1909). The foundations of The origin of species, a sketch written in 1842 (PDF). Cambridge: Printed at the University Press. LCCN 61057537. OCLC 1184581. Archived (PDF) from the original on 4 March 2016. Retrieved 27 November 2014.
- Hall, Brian K.; Hallgrímsson, Benedikt (6 December 2007). Strickberger's Evolution. Jones & Bartlett Publishers. ISBN 978-1-4496-4722-3.
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). Molecular Biology of the Cell (4th ed.). Garland. ISBN 978-0-8153-3218-3. OCLC 145080076.
- Begon M, Townsend CR, Harper JL (2005). Ecology: From Individuals to Ecosystems (4th ed.). Blackwell Publishing Limited. ISBN 978-1-4051-1117-1. OCLC 57639896.
- Campbell N (2004). Biology (7th ed.). Benjamin-Cummings Publishing Company. ISBN 978-0-8053-7146-8. OCLC 71890442.
- Colinvaux P (1979). Why Big Fierce Animals are Rare: An Ecologist's Perspective (reissue ed.). Princeton University Press. ISBN 978-0-691-02364-9. OCLC 10081738.
- Mayr, Ernst (1982). The Growth of Biological Thought: Diversity, Evolution, and Inheritance. Harvard University Press. ISBN 978-0-674-36446-2. Archived from the original on 2015-10-03. Retrieved 2015-06-27.
- Hoagland M (2001). The Way Life Works (reprint ed.). Jones and Bartlett Publishers inc. ISBN 978-0-7637-1688-2. OCLC 223090105.
- Janovy, John (2004). On Becoming a Biologist (2nd ed.). Bison Books. ISBN 978-0-8032-7620-8. OCLC 55138571.
- Johnson, George B. (2005). Biology, Visualizing Life. Holt, Rinehart, and Winston. ISBN 978-0-03-016723-2. OCLC 36306648.
- Tobin, Allan; Dusheck, Jennie (2005). Asking About Life (3rd ed.). Belmont, Calif.: Wadsworth. ISBN 978-0-534-40653-0.
- IPCC (2013). Stocker, T. F.; Qin, D.; Plattner, G.-K.; Tignor, M.; et al. (eds.). Climate Change 2013: The Physical Science Basis (PDF). Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, UK & New York: Cambridge University Press. ISBN 978-1-107-05799-9..
- IPCC (2013). "Summary for Policymakers" (PDF). IPCC AR5 WG1 2013.
- IPCC (2018). Masson-Delmotte, V.; Zhai, P.; Pörtner, H.-O.; Roberts, D.; et al. (eds.). Global Warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty (PDF). Intergovernmental Panel on Climate Change. Global Warming of 1.5 ºC —.
- Allen, M. R.; Dube, O. P.; Solecki, W.; Aragón-Durand, F.; et al. (2018). "Chapter 1: Framing and Context" (PDF). IPCC SR15 2018. pp. 49–91.
- US EPA (15 September 2020). "Overview of Greenhouse Gases". Retrieved 15 September 2020.
External links
- Biology at Curlie
- OSU's Phylocode
- Biology Online – Wiki Dictionary
- MIT video lecture series on biology
- OneZoom Tree of Life
Journal links
- PLOS Biology A peer-reviewed, open-access journal published by the Public Library of Science
- Current Biology: General journal publishing original research from all areas of biology
- Biology Letters: A high-impact Royal Society journal publishing peer-reviewed biology papers of general interest
- Science: Internationally renowned AAAS science journal – see sections of the life sciences
- International Journal of Biological Sciences: A biological journal publishing significant peer-reviewed scientific papers
- Perspectives in Biology and Medicine: An interdisciplinary scholarly journal publishing essays of broad relevance