Chromosome 4
| Chromosome 4 | |
|---|---|
 Human chromosome 4 pair after G-banding. One is from mother, one is from father. | |
 Chromosome 4 pair in human male karyogram. | |
| Features | |
| Length (bp) | 190,214,555 bp (GRCh38)[1] |
| No. of genes | 727 (CCDS)[2] |
| Type | Autosome |
| Centromere position | Submetacentric[3] (50.0 Mbp[4]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome 4 |
| Entrez | Chromosome 4 |
| NCBI | Chromosome 4 |
| UCSC | Chromosome 4 |
| Full DNA sequences | |
| RefSeq | NC_000004 (FASTA) |
| GenBank | CM000666 (FASTA) |
Chromosome 4 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 4 spans more than 186 million base pairs (the building material of DNA) and represents between 6 and 6.5 percent of the total DNA in cells.
Genomics
The chromosome is ~191 megabases in length. In a 2012 paper, 775 protein-encoding genes were identified on this chromosome.[5] 211 (27.9%) of these coding sequences did not have any experimental evidence at the protein level, in 2012. 271 appear to be membrane proteins. 54 have been classified as cancer-associated proteins.
Genes
Number of genes
The following are some of the gene count estimates of human chromosome 4. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[6]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 727 | — | — | [2] | 2016-09-08 |
| HGNC | 731 | 277 | 633 | [7] | 2017-05-12 |
| Ensembl | 746 | 993 | 727 | [8] | 2017-03-29 |
| UniProt | 765 | — | — | [9] | 2018-02-28 |
| NCBI | 769 | 934 | 819 | [10][11][12] | 2017-05-19 |
Gene list
The following is a partial list of genes on human chromosome 4. For complete list, see the link in the infobox on the right.
- AASDH: aminoadipate-semialdehyde dehydrogenase
- ACVR1: activin-like kinase 2 (ALK-2)
- ACOX3: encoding enzyme Peroxisomal acyl-coenzyme A oxidase 3
- AGA: AGU syndrome (Finnish heritage disease) related gene
- AGPAT9: encoding enzyme Glycerol-3-phosphate acyltransferase 3 a.k.a. 1-acylglycerol-3-phosphate O-acyltransferase 9
- ANK2: ankyrin 2, neuronal
- APBB2: encoding protein Amyloid beta A4 precursor protein-binding family B member 2
- ART3: encoding enzyme Ecto-ADP-ribosyltransferase 3
- ASAHL: encoding enzyme N-acylethanolamine-hydrolyzing acid amidase
- FAM198B: encoding protein Protein ENED
- ZGRF1: zinc-finger GRF-type containing 1
- CCDC109B: Coiled-coil domain containing 109B
- Complement Factor I: Complement Factor I
- CRMP1: Collapsin response mediator protein 1, a member of CRMP family
- CSN2: Beta-casein
- CXCL1: chemokine (C-X-C motif) ligand 1, scyb1
- CXCL2: chemokine (C-X-C motif) ligand 2, scyb2
- CXCL3: chemokine (C-X-C motif) ligand 3, scyb3
- CXCL4: chemokine (C-X-C motif) ligand 4, Platelet factor-4, PF-4, scyb4
- CXCL5: chemokine (C-X-C motif) ligand 5, scyb5
- CXCL6: chemokine (C-X-C motif) ligand 6, scyb6
- CXCL7: chemokine (C-X-C motif) ligand 7, PPBP, scyb7
- CXCL8: chemokine (C-X-C motif) ligand 8, interleukin 8 (IL-8), scyb8
- CXCL9: chemokine (C-X-C motif) ligand 9, scyb9
- CXCL10: chemokine (C-X-C motif) ligand 10, scyb10
- CXCL11: chemokine (C-X-C motif) ligand 11, scyb11
- CXCL13: chemokine (C-X-C motif) ligand 13, scyb13
- CYTL1: Cytokine-like 1
- DCUN1D4: Defective in cullin neddylation 1 domain containing 4
- DHX15: DEAH-box helicase 15
- DKK2: Dickkopf-related protein 2
- DUX4: Thought to be inactive but 2010 research shows a key role in FSHD[13]
- ELMOD2: Elmo domain-containing 2
- EMCN: Endomucin
- EVC: Ellis van Creveld syndrome
- EVC2: Ellis van Creveld syndrome 2 (limbin)
- Factor XI: Mutations cause Haemophilia C
- FAM47E-STBD1: FAM47E-STBD1 readthrough
- FAM114A1: Family with sequence similarity 114, member A1
- FAM149A: Family with sequence similarity 149, member A
- FAM193A: Family with sequence similarity 193, member A
- FAM221B: Family with sequence similarity 221, member B
- FGF2: Fibroblast growth factor 2 (basic fibroblast growth factor)
- FGFR3: fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism, bladder cancer)
- FGFRL1: fibroblast growth factor receptor-like 1
- FRG1: FSHD region gene 1
- GUF1: GUF1 homolog, GTPase
- HCL2: encoding hair color 2 (red) protein
- HDL3: encoding Huntington-like neurodegenerative disorder 2 protein
- HTT (Huntingtin): huntingtin protein (Huntington's disease)
- IGJ: linker protein for immunoglobulin alpha and mu polypeptides
- INTS12: Integrator complex subunit 12
- KDR: Kinase insert domain receptor (Vascular endothelial growth factor receptor 2)
- KIAA1530: UV stimulated scaffold protein A
- LCORL: Ligand dependent nuclear receptor corepressor like
- LDB2: LIM domain-binding protein 2
- LGI2: Leucine-rich repeat LGI family member 2
- LOC100505912 encoding protein Uncharacterized LOC100505912
- LSM6: U6 snRNA-associated Sm-like protein
- LYAR: Cell growth-regulating nucleolar protein
- MAB21L2: Mab-21-like 2
- Marcksl1: encoding protein MARCKS-like 1
- MAML3: Mastermind-like 3
- MFSD7: encoding protein Major facilitator superfamily domain containing 7
- MIR1269A: microRNA 1269a
- MLF1IP: Centromere protein U
- MMAA: methylmalonic aciduria (cobalamin deficiency) cblA type
- MTHFD2L: NAD-dependent methylenetetrahydrofolate dehydrogenase 2-like protein
- MYL5: Myosin light chain 5
- NOA1: encoding protein Nitric oxide associated 1
- NUDT6: nudix hydrolase 6
- NUDT9: nudix hydrolase 9
- OTUD4: OTU domain-containing protein 4
- PABPC4L: encoding protein Poly(A) binding protein, cytoplasmic 4-like
- PARM1: Prostate androgen-regulated mucin-like protein 1
- PHOX2B: codes for a homeodomain transcription factor
- PI4K2B: Phosphatidylinositol 4-kinase type 2-beta
- PKD2: polycystic kidney disease 2 (autosomal dominant)
- PLK4: Serine/threonine-protein kinase PLK4
- PSAPL1: encoding protein Prosaposin-like 1 (gene/pseudogene)
- QDPR: quinoid dihydropteridine reductase
- RBM47: RNA binding motif protein 47
- SDAD1: protein SDA1 homolog
- SEC24B: Sec24 homolog B
- SEC24D: Sec24 homolog D
- SEPT11: Septin-11
- SLC9B2: solute carrier family 9 member B2
- SLC10A4: solute carrier family 10 member 4
- SMIM20: encoding protein Small integral membrane protein 20
- SNCA: synuclein, alpha (non A4 component of amyloid precursor)
- SPATA5: Spermatogenesis-associated protein 5
- STATH: gene with protein product
- TACC3: Transforming acidic coiled-coil-containing protein 3
- TENM3: Teneurin transmembrane protein 3
- THAP6: THAP domain-containing protein 6
- TMPRSS11D: Transmembrane protease, serine 11D
- TNIP2: TNFAIP3-interaction protein 2
- UCHL1: ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
- UGT8: UDP glycosyltransferase 8
- UNC5C: netrin receptor UNC5C
- USP38: encoding protein Ubiquitin specific peptidase 38
- USP53: ubiquitin specific peptidase 53
- UTP3: small subunit processome component
- WFS1: Wolfram syndrome 1 (wolframin)
- ZNF621: encoding protein Zinc finger protein 621
Diseases and disorders
The following are some of the diseases related to genes located on chromosome 4:
- Achondroplasia
- Autosomal dominant polycystic kidney disease (PKD-2)
- Bladder cancer
- Crouzonodermoskeletal syndrome
- Chronic lymphocytic leukemia
- Congenital central hypoventilation syndrome
- Ellis-van Creveld syndrome
- Facioscapulohumeral muscular dystrophy
- Fibrodysplasia ossificans progressiva (FOP)
- Haemophilia C
- Huntington's disease
- Hemolytic uremic syndrome
- Hereditary benign intraepithelial dyskeratosis
- Hirschprung's disease
- Hypochondroplasia
- Methylmalonic acidemia
- Mucopolysaccharidosis type I
- Muenke syndrome
- Nonsyndromic deafness
- Nonsyndromic deafness, autosomal dominant
- Parkinson's disease
- Polycystic kidney disease
- Romano-Ward syndrome
- SADDAN
- Tetrahydrobiopterin deficiency
- Thanatophoric dysplasia
- Type 1
- Type 2
- Wolfram syndrome
- Wolf–Hirschhorn syndrome
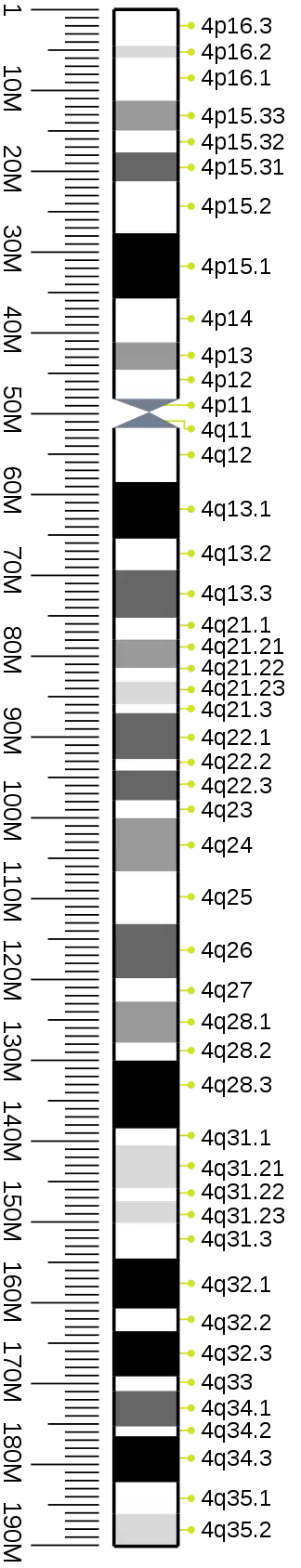
Cytogenetic band
| Chr. | Arm[19] | Band[20] | ISCN start[21] |
ISCN stop[21] |
Basepair start |
Basepair stop |
Stain[22] | Density |
|---|---|---|---|---|---|---|---|---|
| 4 | p | 16.3 | 0 | 220 | 1 | 4500000 | gneg | |
| 4 | p | 16.2 | 220 | 389 | 4,500,001 | 6,000,000 | gpos | 25 |
| 4 | p | 16.1 | 389 | 779 | 6,000,001 | 11,300,000 | gneg | |
| 4 | p | 15.33 | 779 | 1066 | 11,300,001 | 15,000,000 | gpos | 50 |
| 4 | p | 15.32 | 1066 | 1286 | 15,000,001 | 17,700,000 | gneg | |
| 4 | p | 15.31 | 1286 | 1557 | 17,700,001 | 21,300,000 | gpos | 75 |
| 4 | p | 15.2 | 1557 | 1811 | 21,300,001 | 27,700,000 | gneg | |
| 4 | p | 15.1 | 1811 | 2166 | 27,700,001 | 35,800,000 | gpos | 100 |
| 4 | p | 14 | 2166 | 2505 | 35,800,001 | 41,200,000 | gneg | |
| 4 | p | 13 | 2505 | 2742 | 41,200,001 | 44,600,000 | gpos | 50 |
| 4 | p | 12 | 2742 | 2877 | 44,600,001 | 48,200,000 | gneg | |
| 4 | p | 11 | 2877 | 3046 | 48,200,001 | 50,000,000 | acen | |
| 4 | q | 11 | 3046 | 3249 | 50,000,001 | 51,800,000 | acen | |
| 4 | q | 12 | 3249 | 3571 | 51,800,001 | 58,500,000 | gneg | |
| 4 | q | 13.1 | 3571 | 3910 | 58,500,001 | 65,500,000 | gpos | 100 |
| 4 | q | 13.2 | 3910 | 4062 | 65,500,001 | 69,400,000 | gneg | |
| 4 | q | 13.3 | 4062 | 4333 | 69,400,001 | 75,300,000 | gpos | 75 |
| 4 | q | 21.1 | 4333 | 4502 | 75,300,001 | 78,000,000 | gneg | |
| 4 | q | 21.21 | 4502 | 4671 | 78,000,001 | 81,500,000 | gpos | 50 |
| 4 | q | 21.22 | 4671 | 4739 | 81,500,001 | 83,200,000 | gneg | |
| 4 | q | 21.23 | 4739 | 4874 | 83,200,001 | 86,000,000 | gpos | 25 |
| 4 | q | 21.3 | 4874 | 5145 | 86,000,001 | 87,100,000 | gneg | |
| 4 | q | 22.1 | 5145 | 5517 | 87,100,001 | 92,800,000 | gpos | 75 |
| 4 | q | 22.2 | 5517 | 5636 | 92,800,001 | 94,200,000 | gneg | |
| 4 | q | 22.3 | 5636 | 5890 | 94,200,001 | 97,900,000 | gpos | 75 |
| 4 | q | 23 | 5890 | 6059 | 97,900,001 | 100,100,000 | gneg | |
| 4 | q | 24 | 6059 | 6347 | 100,100,001 | 106,700,000 | gpos | 50 |
| 4 | q | 25 | 6347 | 6685 | 106,700,001 | 113,200,000 | gneg | |
| 4 | q | 26 | 6685 | 7040 | 113,200,001 | 119,900,000 | gpos | 75 |
| 4 | q | 27 | 7040 | 7277 | 119,900,001 | 122,800,000 | gneg | |
| 4 | q | 28.1 | 7277 | 7565 | 122,800,001 | 127,900,000 | gpos | 50 |
| 4 | q | 28.2 | 7565 | 7734 | 127,900,001 | 130,100,000 | gneg | |
| 4 | q | 28.3 | 7734 | 8259 | 130,100,001 | 138,500,000 | gpos | 100 |
| 4 | q | 31.1 | 8259 | 8581 | 138,500,001 | 140,600,000 | gneg | |
| 4 | q | 31.21 | 8581 | 8733 | 140,600,001 | 145,900,000 | gpos | 25 |
| 4 | q | 31.22 | 8733 | 8851 | 145,900,001 | 147,500,000 | gneg | |
| 4 | q | 31.23 | 8851 | 9004 | 147,500,001 | 150,200,000 | gpos | 25 |
| 4 | q | 31.3 | 9004 | 9207 | 150,200,001 | 154,600,000 | gneg | |
| 4 | q | 32.1 | 9207 | 9545 | 154,600,001 | 160,800,000 | gpos | 100 |
| 4 | q | 32.2 | 9545 | 9681 | 160,800,001 | 163,600,000 | gneg | |
| 4 | q | 32.3 | 9681 | 9985 | 163,600,001 | 169,200,000 | gpos | 100 |
| 4 | q | 33 | 9985 | 10087 | 169,200,001 | 171,000,000 | gneg | |
| 4 | q | 34.1 | 10087 | 10341 | 171,000,001 | 175,400,000 | gpos | 75 |
| 4 | q | 34.2 | 10341 | 10408 | 175,400,001 | 176,600,000 | gneg | |
| 4 | q | 34.3 | 10408 | 10628 | 176,600,001 | 182,300,000 | gpos | 100 |
| 4 | q | 35.1 | 10628 | 10967 | 182,300,001 | 186,200,000 | gneg | |
| 4 | q | 35.2 | 10967 | 11170 | 186,200,001 | 190,214,555 | gpos | 25 |
References
- ↑ "Human Genome Assembly GRCh38 - Genome Reference Consortium". National Center for Biotechnology Information. 2013-12-24. Retrieved 2017-03-04.
- 1 2 "Search results - 4[CHR] AND "Homo sapiens"[Organism] AND ("has ccds"[Properties] AND alive[prop]) - Gene". NCBI. CCDS Release 20 for Homo sapiens. 2016-09-08. Retrieved 2017-05-28.
- ↑ Tom Strachan; Andrew Read (2 April 2010). Human Molecular Genetics. Garland Science. p. 45. ISBN 978-1-136-84407-2.
- 1 2 Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
- ↑ Chen LC, Liu MY, Hsiao YC, Choong WK, Wu HY, Hsu WL, Liao PC, Sung TY, Tsai SF, Yu JS, Chen YJ (2012) Decoding the disease-associated proteins encoded in the human chromosome 4. J Proteome Res
- ↑ Pertea M, Salzberg SL (2010). "Between a chicken and a grape: estimating the number of human genes". Genome Biol. 11 (5): 206. doi:10.1186/gb-2010-11-5-206. PMC 2898077. PMID 20441615.
- ↑ "Statistics & Downloads for chromosome 4". HUGO Gene Nomenclature Committee. 2017-05-12. Retrieved 2017-05-19.
- ↑ "Chromosome 4: Chromosome summary - Homo sapiens". Ensembl Release 88. 2017-03-29. Retrieved 2017-05-19.
- ↑ "Human chromosome 4: entries, gene names and cross-references to MIM". UniProt. 2018-02-28. Retrieved 2018-03-16.
- ↑ "Search results - 4[CHR] AND "Homo sapiens"[Organism] AND ("genetype protein coding"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ↑ "Search results - 4[CHR] AND "Homo sapiens"[Organism] AND ( ("genetype miscrna"[Properties] OR "genetype ncrna"[Properties] OR "genetype rrna"[Properties] OR "genetype trna"[Properties] OR "genetype scrna"[Properties] OR "genetype snrna"[Properties] OR "genetype snorna"[Properties]) NOT "genetype protein coding"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ↑ "Search results - 4[CHR] AND "Homo sapiens"[Organism] AND ("genetype pseudo"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ↑ Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, Tawil R, Frants RR, van der Maarel SM (September 2010). "A unifying genetic model for facioscapulohumeral muscular dystrophy". Science. 329 (5999): 1650–3. Bibcode:2010Sci...329.1650L. doi:10.1126/science.1189044. PMC 4677822. PMID 20724583.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (400 bphs, Assembly GRCh38.p3). Last update 2014-03-04. Retrieved 2017-04-26.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (550 bphs, Assembly GRCh38.p3). Last update 2015-08-11. Retrieved 2017-04-26.
- ↑ International Standing Committee on Human Cytogenetic Nomenclature (2013). ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Karger Medical and Scientific Publishers. ISBN 978-3-318-02253-7.
- ↑ Sethakulvichai, W.; Manitpornsut, S.; Wiboonrat, M.; Lilakiatsakun, W.; Assawamakin, A.; Tongsima, S. (2012). "Estimation of band level resolutions of human chromosome images". In Computer Science and Software Engineering (JCSSE), 2012 International Joint Conference on: 276–282. doi:10.1109/JCSSE.2012.6261965. ISBN 978-1-4673-1921-8. S2CID 16666470.
- ↑ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
- ↑ "p": Short arm; "q": Long arm.
- ↑ For cytogenetic banding nomenclature, see article locus.
- 1 2 These values (ISCN start/stop) are based on the length of bands/ideograms from the ISCN book, An International System for Human Cytogenetic Nomenclature (2013). Arbitrary unit.
- ↑ gpos: Region which is positively stained by G banding, generally AT-rich and gene poor; gneg: Region which is negatively stained by G banding, generally CG-rich and gene rich; acen Centromere. var: Variable region; stalk: Stalk.
Further reading
- Goldfrank D, Schoenberger E, Gilbert F (2003). "Disease genes and chromosomes: disease maps of the human genome. Chromosome 4". Genet Test. 7 (4): 351–72. doi:10.1089/109065703322783752. PMID 15000816.
- Hillier LW, Graves TA, Fulton RS, Fulton LA, Pepin KH, Minx P, et al. (April 2005). "Generation and annotation of the DNA sequences of human chromosomes 2 and 4". Nature. 434 (7034): 724–31. Bibcode:2005Natur.434..724H. doi:10.1038/nature03466. PMID 15815621.
External links
| Wikimedia Commons has media related to Human chromosome 4. |
- National Institutes of Health. "Chromosome 4". Genetics Home Reference. Archived from the original on August 3, 2004. Retrieved 2017-05-06.
- "Chromosome 4". Human Genome Project Information Archive 1990–2003. Retrieved 2017-05-06.